| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
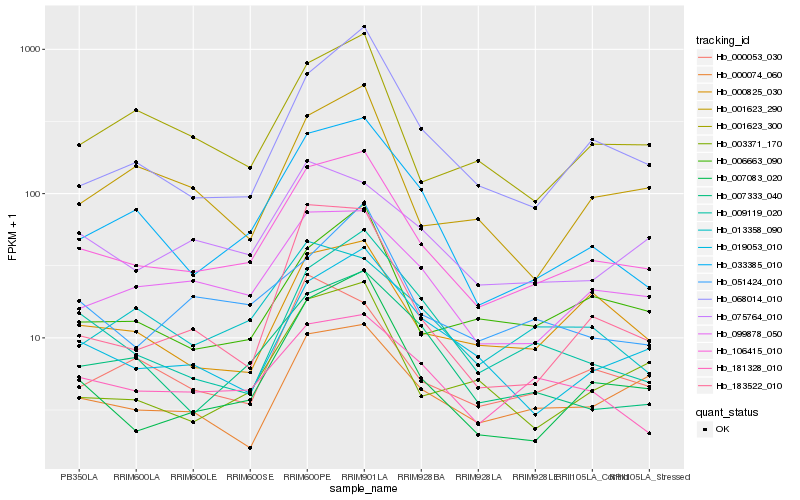
Hb_106415_010 |
0.0 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 2 |
Hb_099878_050 |
0.1098802989 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 3 |
Hb_033385_010 |
0.1160268368 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 4 |
Hb_007083_020 |
0.1288382392 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 5 |
Hb_019053_010 |
0.129048207 |
- |
- |
- |
| 6 |
Hb_007333_040 |
0.1303286237 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 7 |
Hb_000825_030 |
0.1304176914 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_181328_010 |
0.1314187315 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_006663_090 |
0.1319124772 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 10 |
Hb_068014_010 |
0.1343688269 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 11 |
Hb_001623_300 |
0.1353089013 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 12 |
Hb_000074_060 |
0.1374484969 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 13 |
Hb_003371_170 |
0.1376138902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_051424_010 |
0.1403926801 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 15 |
Hb_183522_010 |
0.1440654648 |
- |
- |
Far upstream element-binding protein, putative [Ricinus communis] |
| 16 |
Hb_000053_030 |
0.1442368944 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 17 |
Hb_009119_020 |
0.1445210003 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 18 |
Hb_001623_290 |
0.1450757242 |
- |
- |
unknown [Populus trichocarpa] |
| 19 |
Hb_075764_010 |
0.1465545318 |
- |
- |
PREDICTED: PRA1 family protein A1-like [Jatropha curcas] |
| 20 |
Hb_013358_090 |
0.148743109 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |