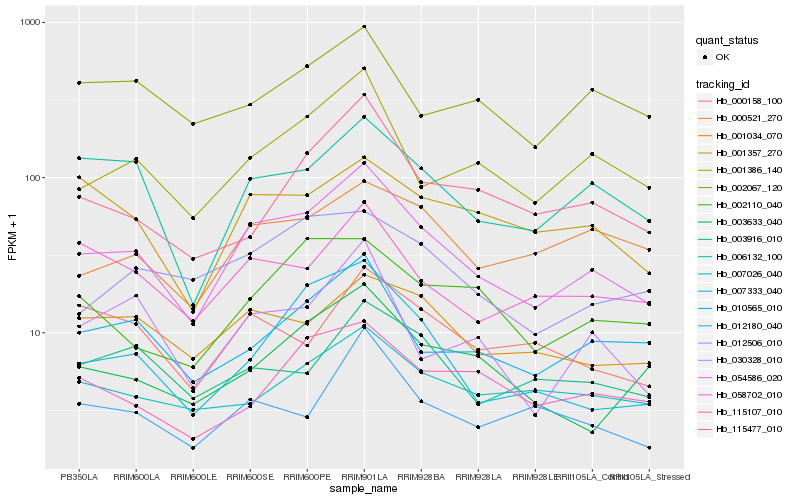
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_054586_020 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 2 |
Hb_001386_140 |
0.1325692949 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 3 |
Hb_006132_100 |
0.1329800822 |
- |
- |
PREDICTED: uncharacterized protein LOC105642968 [Jatropha curcas] |
| 4 |
Hb_010565_010 |
0.1387687619 |
- |
- |
PREDICTED: N-acetylglucosaminyl-phosphatidylinositol de-N-acetylase-like isoform X2 [Populus euphratica] |
| 5 |
Hb_058702_010 |
0.1433310812 |
- |
- |
PREDICTED: importin beta-like SAD2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007333_040 |
0.1447907527 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 7 |
Hb_030328_010 |
0.1456386578 |
- |
- |
hypothetical protein CICLE_v10015936mg [Citrus clementina] |
| 8 |
Hb_001034_070 |
0.1458663412 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_012180_040 |
0.1478351211 |
- |
- |
hypothetical protein VITISV_042517 [Vitis vinifera] |
| 10 |
Hb_003633_040 |
0.1519587769 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 11 |
Hb_115477_010 |
0.1521230855 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 12 |
Hb_003916_010 |
0.1536621233 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 13 [Jatropha curcas] |
| 13 |
Hb_012506_010 |
0.1536989502 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 14 |
Hb_000521_270 |
0.1540536457 |
- |
- |
PREDICTED: protein EMSY-LIKE 3 [Jatropha curcas] |
| 15 |
Hb_001357_270 |
0.1541554412 |
- |
- |
PREDICTED: aspartic proteinase A1-like [Jatropha curcas] |
| 16 |
Hb_000158_100 |
0.1547336199 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_007026_040 |
0.1549447264 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |
| 18 |
Hb_002110_040 |
0.1582322243 |
- |
- |
hypothetical protein AMTR_s00135p00074190 [Amborella trichopoda] |
| 19 |
Hb_115107_010 |
0.1599878121 |
- |
- |
Early-responsive to dehydration stress protein (ERD4) isoform 1 [Theobroma cacao] |
| 20 |
Hb_002067_120 |
0.1603814636 |
- |
- |
PREDICTED: uncharacterized protein LOC105111554 isoform X1 [Populus euphratica] |