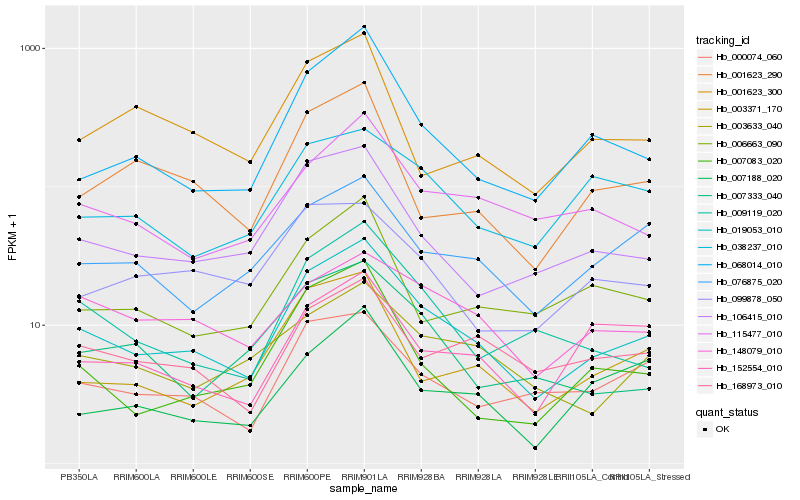
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019053_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_068014_010 |
0.1287800097 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 3 |
Hb_106415_010 |
0.129048207 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 4 |
Hb_009119_020 |
0.1373925548 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 5 |
Hb_000074_060 |
0.140185695 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 6 |
Hb_115477_010 |
0.1443285789 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 7 |
Hb_003633_040 |
0.1469036622 |
- |
- |
hypothetical protein B456_008G146200 [Gossypium raimondii] |
| 8 |
Hb_003371_170 |
0.1487285226 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_168973_010 |
0.1493327235 |
- |
- |
- |
| 10 |
Hb_076875_020 |
0.1516113686 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 17B [Jatropha curcas] |
| 11 |
Hb_007083_020 |
0.1557941862 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 12 |
Hb_148079_010 |
0.1563932072 |
- |
- |
PREDICTED: WD repeat-containing protein 82 [Jatropha curcas] |
| 13 |
Hb_001623_290 |
0.1566341886 |
- |
- |
unknown [Populus trichocarpa] |
| 14 |
Hb_006663_090 |
0.1581151375 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 15 |
Hb_007333_040 |
0.1585889303 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 16 |
Hb_099878_050 |
0.1615443983 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 17 |
Hb_152554_010 |
0.1617232904 |
- |
- |
hypothetical protein B456_008G095800, partial [Gossypium raimondii] |
| 18 |
Hb_038237_010 |
0.1628301392 |
- |
- |
hypothetical protein POPTR_0003s01110g [Populus trichocarpa] |
| 19 |
Hb_007188_020 |
0.1633031795 |
- |
- |
hypothetical protein PRUPE_ppb010436mg [Prunus persica] |
| 20 |
Hb_001623_300 |
0.1680812382 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |