| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
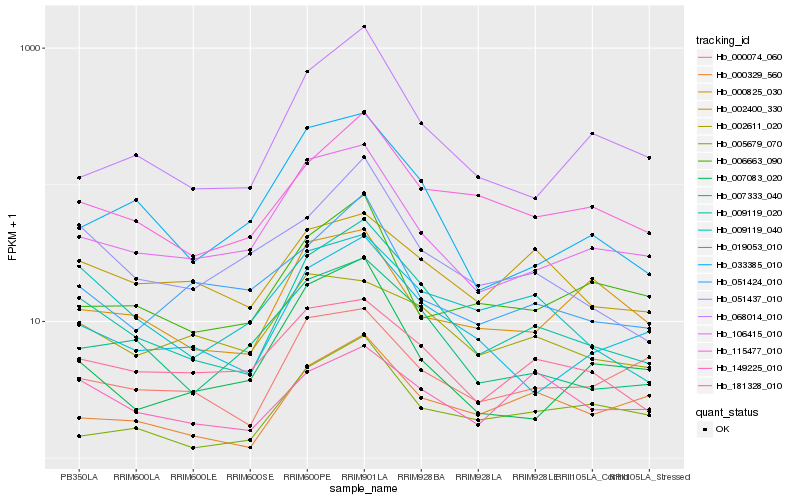
Hb_009119_020 |
0.0 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 2 |
Hb_007333_040 |
0.1331629974 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK1-like [Glycine max] |
| 3 |
Hb_019053_010 |
0.1373925548 |
- |
- |
- |
| 4 |
Hb_115477_010 |
0.1444870223 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 5 |
Hb_106415_010 |
0.1445210003 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 6 |
Hb_051437_010 |
0.1486531834 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 7 |
Hb_033385_010 |
0.1559611942 |
- |
- |
hypothetical protein JCGZ_06094 [Jatropha curcas] |
| 8 |
Hb_068014_010 |
0.1577950954 |
- |
- |
hypothetical protein POPTR_0312s00200g, partial [Populus trichocarpa] |
| 9 |
Hb_181328_010 |
0.1587077478 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 10 |
Hb_009119_040 |
0.1633534466 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2A [Jatropha curcas] |
| 11 |
Hb_000329_560 |
0.173727478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_051424_010 |
0.1741729353 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 13 |
Hb_000074_060 |
0.1795090211 |
- |
- |
PREDICTED: membrane-associated progesterone-binding protein 4-like [Populus euphratica] |
| 14 |
Hb_006663_090 |
0.1843655169 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 15 |
Hb_149225_010 |
0.1848392732 |
- |
- |
DNA-directed RNA polymerase II subunit rpb4, putative isoform 4, partial [Theobroma cacao] |
| 16 |
Hb_007083_020 |
0.1855621908 |
- |
- |
hypothetical protein F775_15403 [Aegilops tauschii] |
| 17 |
Hb_005679_070 |
0.1865689193 |
- |
- |
hypothetical protein M569_07741, partial [Genlisea aurea] |
| 18 |
Hb_002611_020 |
0.1870414143 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 19 |
Hb_002400_330 |
0.1878010868 |
- |
- |
PREDICTED: ER membrane protein complex subunit 7 homolog isoform X2 [Jatropha curcas] |
| 20 |
Hb_000825_030 |
0.1880409217 |
- |
- |
zinc binding dehydrogenase, putative [Ricinus communis] |