| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
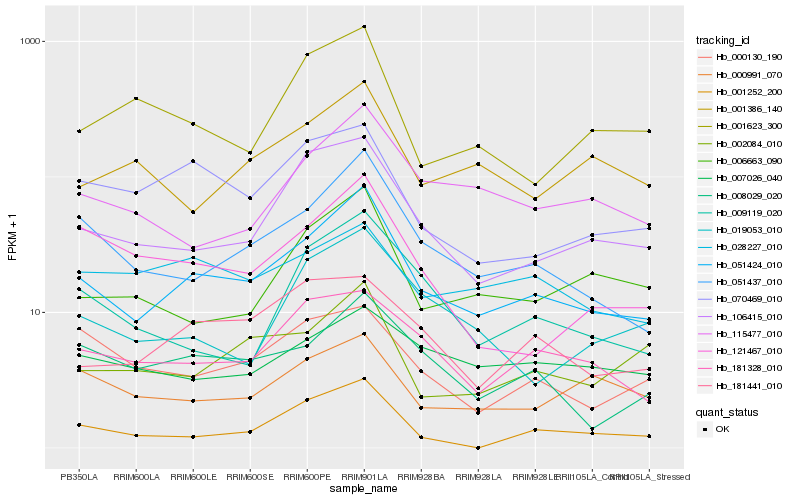
Hb_051424_010 |
0.0 |
- |
- |
PREDICTED: putative methyltransferase At1g22800 [Jatropha curcas] |
| 2 |
Hb_051437_010 |
0.1219084305 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 3 |
Hb_106415_010 |
0.1403926801 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 4 |
Hb_008029_020 |
0.1540060327 |
- |
- |
PREDICTED: uncharacterized protein LOC105631777 [Jatropha curcas] |
| 5 |
Hb_006663_090 |
0.1592067419 |
- |
- |
hypothetical protein B456_010G1255001, partial [Gossypium raimondii] |
| 6 |
Hb_181328_010 |
0.1609541163 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_115477_010 |
0.1611394219 |
- |
- |
Protein dom-3, putative [Ricinus communis] |
| 8 |
Hb_002084_010 |
0.1664256365 |
- |
- |
hypothetical protein VITISV_017990 [Vitis vinifera] |
| 9 |
Hb_181441_010 |
0.1704435774 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 10 |
Hb_070469_010 |
0.1709908547 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 11 |
Hb_121467_010 |
0.1739518012 |
- |
- |
tubulin-specific chaperone E, putative [Ricinus communis] |
| 12 |
Hb_009119_020 |
0.1741729353 |
- |
- |
hypothetical protein CISIN_1g033708mg [Citrus sinensis] |
| 13 |
Hb_019053_010 |
0.1745348089 |
- |
- |
- |
| 14 |
Hb_001386_140 |
0.1788063064 |
- |
- |
PREDICTED: B2 protein [Jatropha curcas] |
| 15 |
Hb_000991_070 |
0.1791911986 |
- |
- |
phytochrome A specific signal transduction component family protein [Populus trichocarpa] |
| 16 |
Hb_001623_300 |
0.1795499398 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 alpha subcomplex subunit 13-B [Jatropha curcas] |
| 17 |
Hb_001252_200 |
0.1821358433 |
- |
- |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 18 |
Hb_000130_190 |
0.1847117043 |
- |
- |
PREDICTED: uncharacterized protein LOC105638437 [Jatropha curcas] |
| 19 |
Hb_028227_010 |
0.1854879226 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 20 |
Hb_007026_040 |
0.1858170627 |
- |
- |
PREDICTED: chaperonin 60 subunit alpha 2, chloroplastic-like [Eucalyptus grandis] |