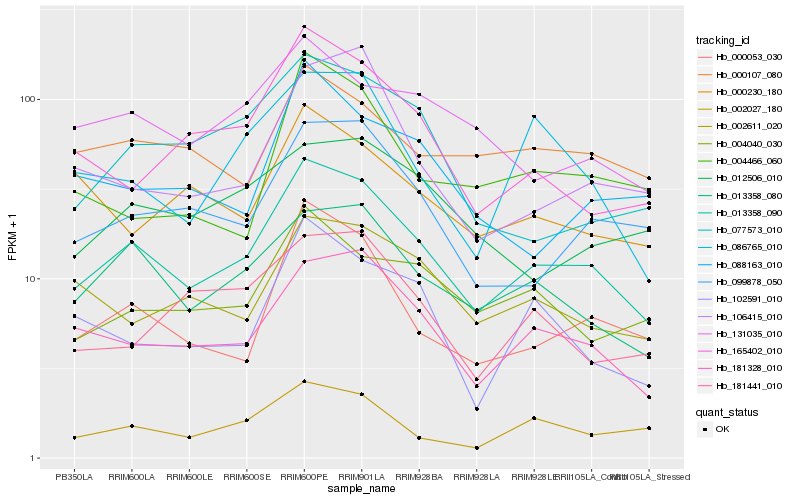
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013358_090 |
0.0 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 2 |
Hb_181328_010 |
0.1083180688 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_013358_080 |
0.1093322892 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 4 |
Hb_099878_050 |
0.1285497652 |
- |
- |
PREDICTED: uncharacterized protein LOC105123495 isoform X2 [Populus euphratica] |
| 5 |
Hb_000053_030 |
0.1292330041 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 6 |
Hb_002027_180 |
0.1364009667 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 7 |
Hb_004040_030 |
0.1393486966 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 8 |
Hb_002611_020 |
0.1415880747 |
- |
- |
PREDICTED: alpha-1,3-mannosyl-glycoprotein 2-beta-N-acetylglucosaminyltransferase-like [Populus euphratica] |
| 9 |
Hb_165402_010 |
0.1419593199 |
- |
- |
PREDICTED: cullin-1 isoform X3 [Nicotiana sylvestris] |
| 10 |
Hb_086765_010 |
0.1460324142 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 11 |
Hb_131035_010 |
0.1473620873 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_102591_010 |
0.1478112827 |
- |
- |
hypothetical protein JCGZ_09876 [Jatropha curcas] |
| 13 |
Hb_106415_010 |
0.148743109 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X1 [Jatropha curcas] |
| 14 |
Hb_000107_080 |
0.1487481731 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 15 |
Hb_077573_010 |
0.1529742823 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 16 |
Hb_181441_010 |
0.1543487754 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 17 |
Hb_012506_010 |
0.1546420282 |
- |
- |
PREDICTED: UDP-D-apiose/UDP-D-xylose synthase 2 [Jatropha curcas] |
| 18 |
Hb_088163_010 |
0.1549515974 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 19 |
Hb_000230_180 |
0.1582630202 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 20 |
Hb_004466_060 |
0.1587155547 |
- |
- |
hypothetical protein POPTR_0009s12110g [Populus trichocarpa] |