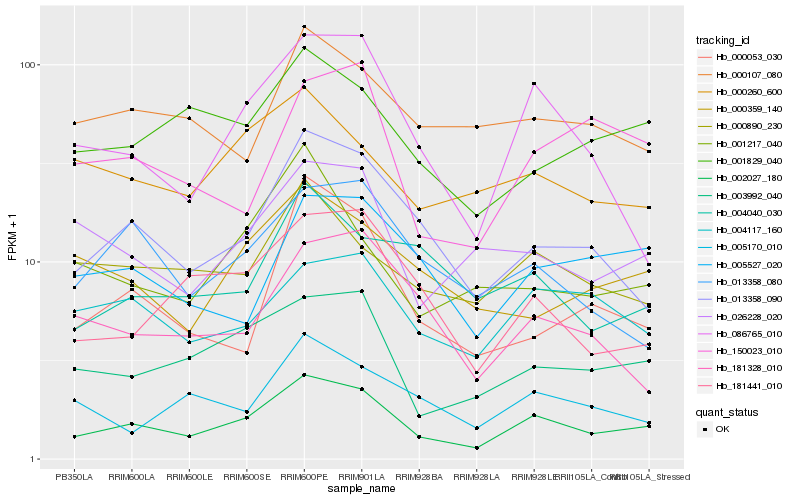
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002027_180 |
0.0 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 2 |
Hb_003992_040 |
0.1323792408 |
- |
- |
Post-illumination chlorophyll fluorescence increase isoform 5 [Theobroma cacao] |
| 3 |
Hb_013358_090 |
0.1364009667 |
- |
- |
hypothetical protein Csa_4G652025 [Cucumis sativus] |
| 4 |
Hb_086765_010 |
0.1489580093 |
- |
- |
PREDICTED: nitrile-specifier protein 5-like [Jatropha curcas] |
| 5 |
Hb_026228_020 |
0.1559492607 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 6 |
Hb_001829_040 |
0.1575689837 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 7 |
Hb_004117_160 |
0.1585923317 |
transcription factor |
TF Family: B3 |
B3 domain-containing transcription repressor VAL2 -like protein [Gossypium arboreum] |
| 8 |
Hb_000890_230 |
0.15861144 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 9 |
Hb_181441_010 |
0.1616605782 |
- |
- |
PREDICTED: uncharacterized protein LOC105640851 [Jatropha curcas] |
| 10 |
Hb_005527_020 |
0.1646479794 |
- |
- |
PREDICTED: uncharacterized protein LOC105643027 [Jatropha curcas] |
| 11 |
Hb_013358_080 |
0.166612273 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 25-like [Jatropha curcas] |
| 12 |
Hb_000359_140 |
0.1675550789 |
- |
- |
PREDICTED: transmembrane protein 56-like [Jatropha curcas] |
| 13 |
Hb_000260_600 |
0.1682984871 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_150023_010 |
0.1713997495 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 15 |
Hb_004040_030 |
0.1720190404 |
- |
- |
PREDICTED: probable inactive purple acid phosphatase 28 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000053_030 |
0.1730672338 |
- |
- |
hypothetical protein CISIN_1g0010393mg, partial [Citrus sinensis] |
| 17 |
Hb_000107_080 |
0.1743589203 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 18 |
Hb_001217_040 |
0.1748015202 |
- |
- |
hypothetical protein MIMGU_mgv1a0002421mg, partial [Erythranthe guttata] |
| 19 |
Hb_005170_010 |
0.175744348 |
- |
- |
- |
| 20 |
Hb_181328_010 |
0.1771405943 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |