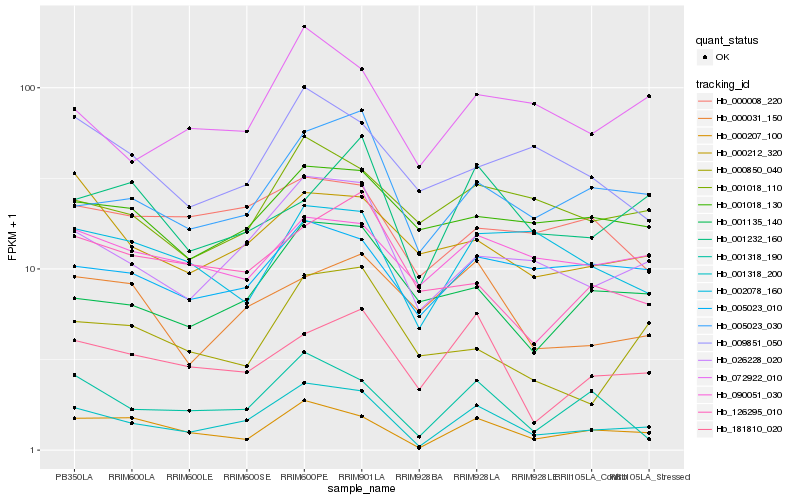
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001318_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 2 |
Hb_026228_020 |
0.1294512773 |
- |
- |
PREDICTED: uncharacterized protein LOC105632747 [Jatropha curcas] |
| 3 |
Hb_000207_100 |
0.1357361685 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 4 |
Hb_005023_030 |
0.1613006515 |
- |
- |
PREDICTED: uncharacterized protein LOC103417254 [Malus domestica] |
| 5 |
Hb_001318_190 |
0.1623914613 |
- |
- |
PREDICTED: uncharacterized protein LOC102617255 [Citrus sinensis] |
| 6 |
Hb_181810_020 |
0.1728534818 |
- |
- |
- |
| 7 |
Hb_001135_140 |
0.1779775842 |
- |
- |
PREDICTED: serine/threonine-protein kinase PBS1-like [Malus domestica] |
| 8 |
Hb_072922_010 |
0.1784138069 |
- |
- |
apolipoprotein d, putative [Ricinus communis] |
| 9 |
Hb_005023_010 |
0.1873952369 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 10 |
Hb_001018_110 |
0.191200757 |
- |
- |
unnamed protein product [Coffea canephora] |
| 11 |
Hb_002078_160 |
0.1917015292 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 12 |
Hb_000212_320 |
0.1953081837 |
- |
- |
PREDICTED: uncharacterized protein LOC105644891 [Jatropha curcas] |
| 13 |
Hb_009851_050 |
0.1969231738 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 14 |
Hb_001232_160 |
0.197801584 |
- |
- |
PREDICTED: protein SYS1 homolog [Gossypium raimondii] |
| 15 |
Hb_000850_040 |
0.1980574036 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_126295_010 |
0.1981753166 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 17 |
Hb_000031_150 |
0.1987661319 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 18 |
Hb_000008_220 |
0.1994387965 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 10 [Jatropha curcas] |
| 19 |
Hb_001018_130 |
0.2001764063 |
- |
- |
PREDICTED: uncharacterized protein LOC105638260 [Jatropha curcas] |
| 20 |
Hb_090051_030 |
0.2007233594 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |