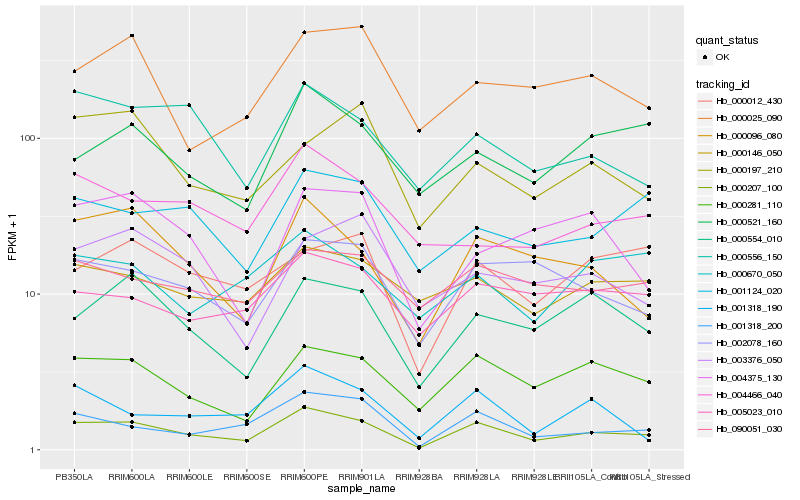
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000207_100 |
0.0 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 2 |
Hb_001318_200 |
0.1357361685 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 3 |
Hb_000281_110 |
0.1388499563 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 4 |
Hb_000096_080 |
0.1399472091 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 5 |
Hb_000554_010 |
0.1525627463 |
- |
- |
transporter, putative [Ricinus communis] |
| 6 |
Hb_002078_160 |
0.1603979246 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 7 |
Hb_000556_150 |
0.1636663514 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 8 |
Hb_001318_190 |
0.1677638951 |
- |
- |
PREDICTED: uncharacterized protein LOC102617255 [Citrus sinensis] |
| 9 |
Hb_000521_160 |
0.1699292478 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001124_020 |
0.1714463578 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 11 |
Hb_000012_430 |
0.1732642648 |
- |
- |
calcineurin B, putative [Ricinus communis] |
| 12 |
Hb_000025_090 |
0.1748237499 |
- |
- |
tubulin beta chain, putative [Ricinus communis] |
| 13 |
Hb_004375_130 |
0.1753087692 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 14 |
Hb_005023_010 |
0.178739149 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 15 |
Hb_000146_050 |
0.1797689273 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |
| 16 |
Hb_004466_040 |
0.1806587335 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 17 |
Hb_090051_030 |
0.1825991381 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 18 |
Hb_000197_210 |
0.1835947288 |
- |
- |
phosphatidylinositol transporter, putative [Ricinus communis] |
| 19 |
Hb_003376_050 |
0.1860900395 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 20 |
Hb_000670_050 |
0.1864628715 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_16088 [Jatropha curcas] |