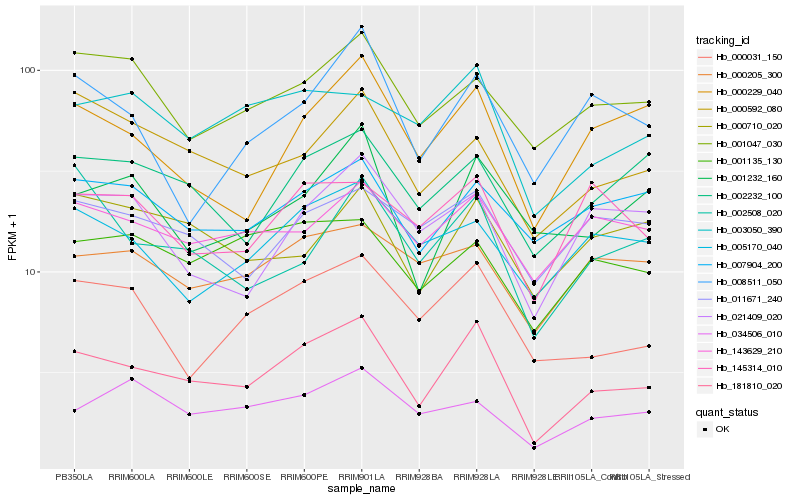
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_181810_020 |
0.0 |
- |
- |
- |
| 2 |
Hb_000031_150 |
0.1294117576 |
- |
- |
PREDICTED: exocyst complex component EXO70A1 [Jatropha curcas] |
| 3 |
Hb_003050_390 |
0.1311341117 |
- |
- |
PREDICTED: uncharacterized protein LOC104442543 [Eucalyptus grandis] |
| 4 |
Hb_002232_100 |
0.1370354197 |
- |
- |
vacuolar ATPase F subunit [Hevea brasiliensis] |
| 5 |
Hb_000229_040 |
0.138190099 |
- |
- |
Secretory carrier membrane protein (SCAMP) family protein isoform 2 [Theobroma cacao] |
| 6 |
Hb_000592_080 |
0.1388462036 |
- |
- |
PREDICTED: thymidylate kinase [Jatropha curcas] |
| 7 |
Hb_143629_210 |
0.1418456609 |
- |
- |
PREDICTED: transcription factor PIF3 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001135_130 |
0.1425234684 |
- |
- |
hypothetical protein CISIN_1g0171752mg, partial [Citrus sinensis] |
| 9 |
Hb_001047_030 |
0.1441428798 |
- |
- |
PREDICTED: protein transport protein SEC13 homolog B [Jatropha curcas] |
| 10 |
Hb_001232_160 |
0.1457382474 |
- |
- |
PREDICTED: protein SYS1 homolog [Gossypium raimondii] |
| 11 |
Hb_005170_040 |
0.1460078492 |
- |
- |
PREDICTED: probable protein S-acyltransferase 7 [Jatropha curcas] |
| 12 |
Hb_145314_010 |
0.1462492554 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 5b-like [Jatropha curcas] |
| 13 |
Hb_007904_200 |
0.1466577971 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 14 |
Hb_000205_300 |
0.1475276239 |
- |
- |
PREDICTED: uncharacterized protein LOC105647767 [Jatropha curcas] |
| 15 |
Hb_011671_240 |
0.1477495489 |
- |
- |
PREDICTED: WD repeat-containing protein WRAP73-like [Jatropha curcas] |
| 16 |
Hb_021409_020 |
0.1479356638 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Malus domestica] |
| 17 |
Hb_034506_010 |
0.147945727 |
- |
- |
PREDICTED: protein OS-9 homolog [Jatropha curcas] |
| 18 |
Hb_000710_020 |
0.1494494696 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MARCH2 isoform X2 [Populus euphratica] |
| 19 |
Hb_002508_020 |
0.1505001583 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit beta-1 [Jatropha curcas] |
| 20 |
Hb_008511_050 |
0.1516701315 |
- |
- |
PREDICTED: cyclase-associated protein 1 [Jatropha curcas] |