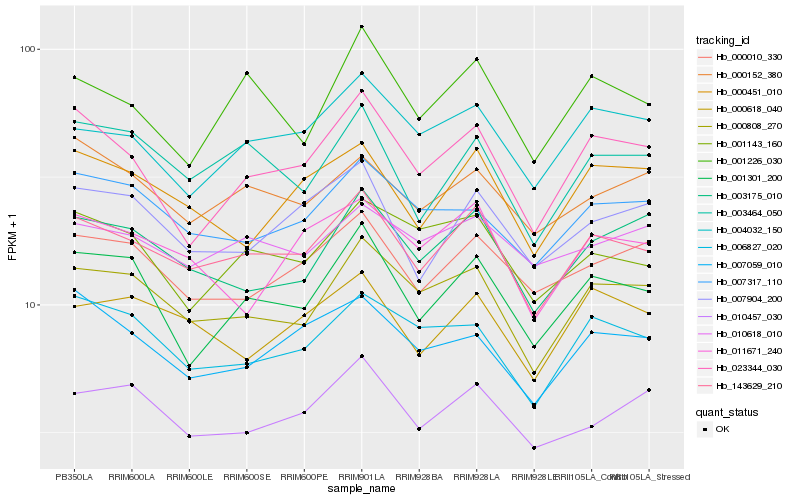
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_143629_210 |
0.0 |
- |
- |
PREDICTED: transcription factor PIF3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000808_270 |
0.0560004129 |
- |
- |
PREDICTED: lon protease homolog 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_003464_050 |
0.0676060936 |
- |
- |
hypothetical protein POPTR_0006s11880g [Populus trichocarpa] |
| 4 |
Hb_004032_150 |
0.0692427007 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 5 |
Hb_000618_040 |
0.0701464534 |
- |
- |
hypothetical protein CICLE_v10027942mg [Citrus clementina] |
| 6 |
Hb_000010_330 |
0.07080827 |
- |
- |
PREDICTED: probable Ufm1-specific protease [Jatropha curcas] |
| 7 |
Hb_000451_010 |
0.0708230033 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 8 |
Hb_001301_200 |
0.0713343307 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 9 |
Hb_003175_010 |
0.0715827666 |
- |
- |
PREDICTED: 39S ribosomal protein L4, mitochondrial [Jatropha curcas] |
| 10 |
Hb_007904_200 |
0.0720329155 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 11 |
Hb_023344_030 |
0.0722608094 |
- |
- |
PREDICTED: putative G3BP-like protein [Jatropha curcas] |
| 12 |
Hb_006827_020 |
0.0729532518 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000152_380 |
0.0741962584 |
- |
- |
PREDICTED: hypersensitive-induced response protein 4 [Jatropha curcas] |
| 14 |
Hb_010457_030 |
0.0742904727 |
- |
- |
APO protein 4, mitochondrial precursor, putative [Ricinus communis] |
| 15 |
Hb_001226_030 |
0.0752741929 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 4 [Jatropha curcas] |
| 16 |
Hb_007317_110 |
0.0757754987 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C660.15-like [Jatropha curcas] |
| 17 |
Hb_010618_010 |
0.0758574095 |
- |
- |
PREDICTED: inactive poly [ADP-ribose] polymerase RCD1 [Jatropha curcas] |
| 18 |
Hb_011671_240 |
0.0761365782 |
- |
- |
PREDICTED: WD repeat-containing protein WRAP73-like [Jatropha curcas] |
| 19 |
Hb_001143_160 |
0.0762193255 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 26 isoform X2 [Jatropha curcas] |
| 20 |
Hb_007059_010 |
0.076294511 |
- |
- |
PREDICTED: uncharacterized protein LOC105642418 [Jatropha curcas] |