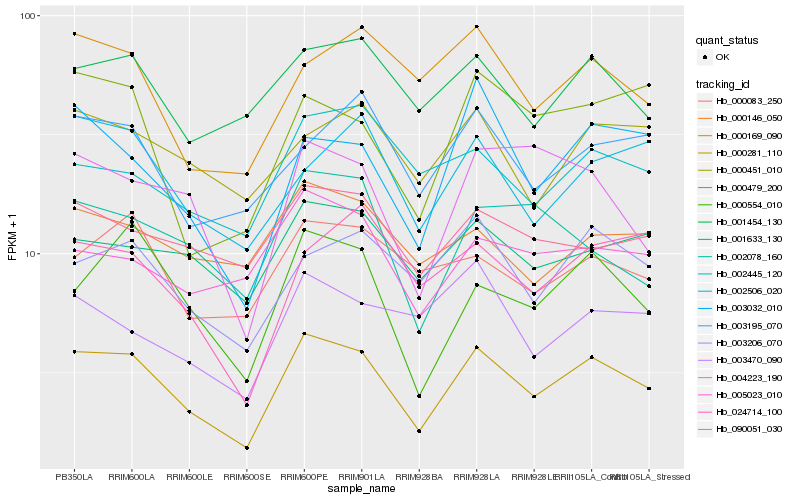
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000281_110 |
0.0 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 2 |
Hb_000554_010 |
0.1108223689 |
- |
- |
transporter, putative [Ricinus communis] |
| 3 |
Hb_003206_070 |
0.1175635897 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 4 |
Hb_003032_010 |
0.1177470766 |
- |
- |
PREDICTED: probable inactive receptor kinase At2g26730 [Jatropha curcas] |
| 5 |
Hb_001454_130 |
0.1201589376 |
- |
- |
Xyloglucan 6-xylosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_001633_130 |
0.1233479873 |
- |
- |
PREDICTED: uncharacterized protein LOC105630200 [Jatropha curcas] |
| 7 |
Hb_000169_090 |
0.123953077 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 8 |
Hb_003195_070 |
0.125761673 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 9 |
Hb_002506_020 |
0.1259784787 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP21-1-like isoform X1 [Populus euphratica] |
| 10 |
Hb_000083_250 |
0.1264297362 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 11 |
Hb_090051_030 |
0.1271419664 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 12 |
Hb_004223_190 |
0.1279238524 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 13 |
Hb_000479_200 |
0.1287162339 |
- |
- |
PREDICTED: uncharacterized protein LOC105644128 [Jatropha curcas] |
| 14 |
Hb_000451_010 |
0.1289772487 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 15 |
Hb_005023_010 |
0.1294024584 |
- |
- |
PREDICTED: putative lipase ROG1 [Jatropha curcas] |
| 16 |
Hb_000146_050 |
0.1294941448 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |
| 17 |
Hb_002078_160 |
0.1296803361 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 18 |
Hb_003470_090 |
0.1314566807 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 19 |
Hb_002445_120 |
0.1315504927 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 20 |
Hb_024714_100 |
0.1316128001 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 isoform X1 [Jatropha curcas] |