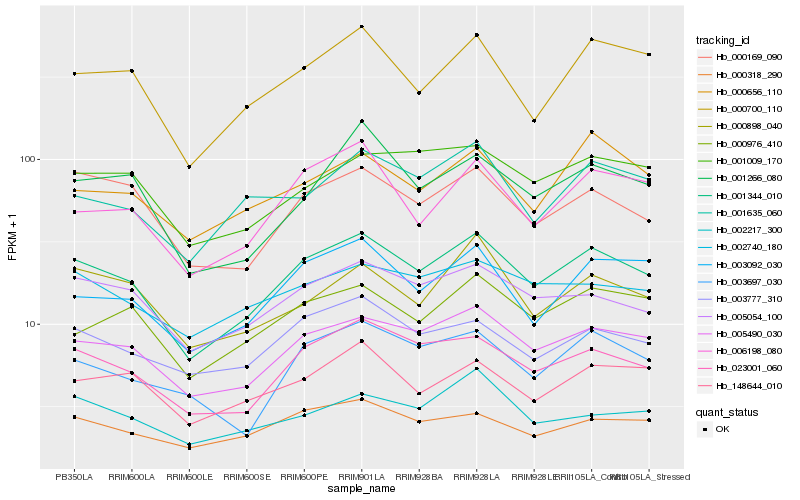
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001344_010 |
0.0 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI11 [Jatropha curcas] |
| 2 |
Hb_005490_030 |
0.0565968093 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 2 isoform X3 [Jatropha curcas] |
| 3 |
Hb_023001_060 |
0.0645615414 |
- |
- |
PREDICTED: importin-11 [Jatropha curcas] |
| 4 |
Hb_005054_100 |
0.0801165569 |
- |
- |
PREDICTED: uncharacterized protein At2g39920 [Jatropha curcas] |
| 5 |
Hb_000169_090 |
0.0828127293 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 6 |
Hb_000700_110 |
0.0837247832 |
- |
- |
Vesicle-associated membrane protein, putative [Ricinus communis] |
| 7 |
Hb_003777_310 |
0.0860253574 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003092_030 |
0.0879902948 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 3 [Jatropha curcas] |
| 9 |
Hb_000976_410 |
0.0888333509 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 10 |
Hb_148644_010 |
0.091022742 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_006198_080 |
0.0921587067 |
- |
- |
hypothetical protein MIMGU_mgv1a013452mg [Erythranthe guttata] |
| 12 |
Hb_001009_170 |
0.0927262217 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 13 |
Hb_002740_180 |
0.0935696023 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000318_290 |
0.093578565 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor VRN1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001635_060 |
0.0942139449 |
- |
- |
eukaryotic translation initiation factor 3 subunit, putative [Ricinus communis] |
| 16 |
Hb_001266_080 |
0.0959727753 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit gamma-like [Jatropha curcas] |
| 17 |
Hb_000898_040 |
0.0960125199 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003697_030 |
0.0963123608 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ATXR2 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000656_110 |
0.0976284947 |
- |
- |
nucleolysin tia-1, putative [Ricinus communis] |
| 20 |
Hb_002217_300 |
0.0991039964 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g48810 [Jatropha curcas] |