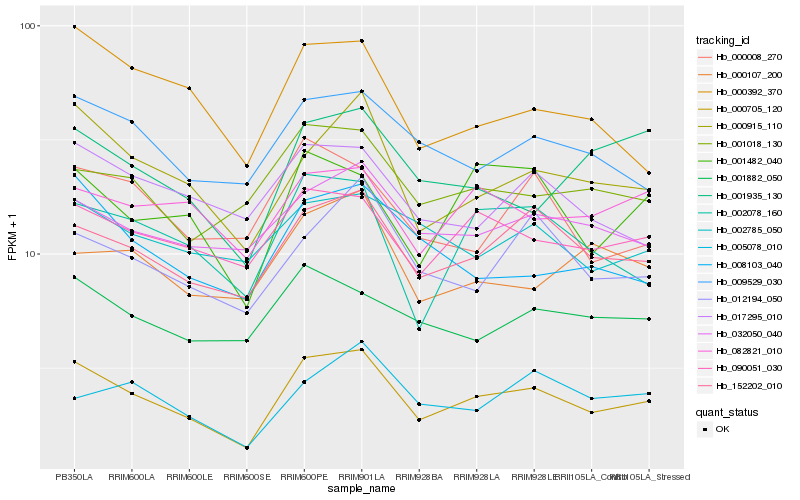
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000705_120 |
0.0 |
- |
- |
glycosyltransferase, putative [Ricinus communis] |
| 2 |
Hb_152202_010 |
0.090104377 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 3 |
Hb_000915_110 |
0.096016538 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase A5 [Jatropha curcas] |
| 4 |
Hb_090051_030 |
0.107114054 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 5 |
Hb_002078_160 |
0.1079337922 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 6 |
Hb_009529_030 |
0.1104659095 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 7 |
Hb_001482_040 |
0.1121652675 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_032050_040 |
0.1142554626 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 9 |
Hb_012194_050 |
0.1159138434 |
- |
- |
Aspartyl aminopeptidase, putative [Ricinus communis] |
| 10 |
Hb_008103_040 |
0.1176769257 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |
| 11 |
Hb_001018_130 |
0.1195488675 |
- |
- |
PREDICTED: uncharacterized protein LOC105638260 [Jatropha curcas] |
| 12 |
Hb_000392_370 |
0.1207776925 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 13 |
Hb_001882_050 |
0.1208873928 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_017295_010 |
0.1215524037 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 15 |
Hb_082821_010 |
0.1230119251 |
- |
- |
PREDICTED: dephospho-CoA kinase-like [Jatropha curcas] |
| 16 |
Hb_005078_010 |
0.1230678816 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 17 |
Hb_001935_130 |
0.1232867785 |
- |
- |
PREDICTED: uncharacterized protein LOC105643176 [Jatropha curcas] |
| 18 |
Hb_000008_270 |
0.1241041633 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein REVOLUTA [Jatropha curcas] |
| 19 |
Hb_002785_050 |
0.1258354138 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 20 |
Hb_000107_200 |
0.1258851655 |
- |
- |
hypothetical protein B456_005G214800 [Gossypium raimondii] |