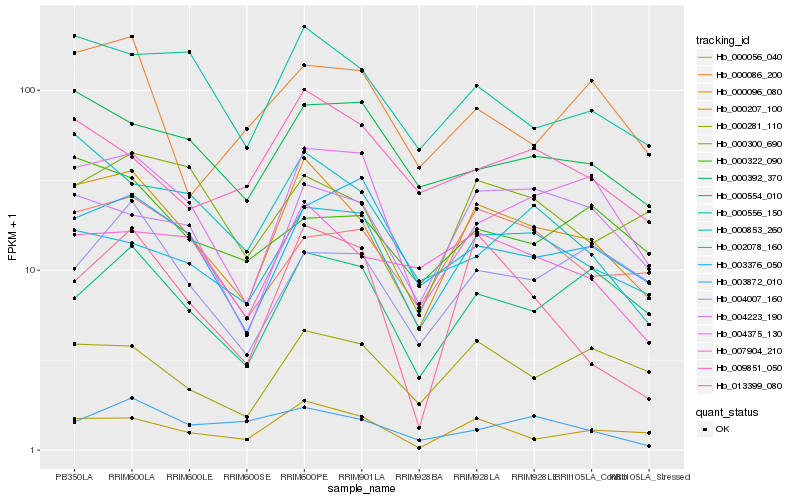
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000096_080 |
0.0 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |
| 2 |
Hb_000556_150 |
0.1324389083 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 3 |
Hb_004375_130 |
0.13750117 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 4 |
Hb_000207_100 |
0.1399472091 |
- |
- |
PREDICTED: ABC transporter G family member 25 [Jatropha curcas] |
| 5 |
Hb_002078_160 |
0.1427650452 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 6 |
Hb_007904_210 |
0.1438367932 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1D [Jatropha curcas] |
| 7 |
Hb_000554_010 |
0.1521543198 |
- |
- |
transporter, putative [Ricinus communis] |
| 8 |
Hb_000281_110 |
0.1543933999 |
- |
- |
PREDICTED: pumilio homolog 12 [Jatropha curcas] |
| 9 |
Hb_004223_190 |
0.1554924354 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 10 |
Hb_000056_040 |
0.1567428307 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31 [Jatropha curcas] |
| 11 |
Hb_000300_690 |
0.1620051845 |
- |
- |
hypothetical protein JCGZ_02719 [Jatropha curcas] |
| 12 |
Hb_000392_370 |
0.164892774 |
- |
- |
PREDICTED: protein IQ-DOMAIN 31-like [Jatropha curcas] |
| 13 |
Hb_009851_050 |
0.172791229 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 14 |
Hb_003376_050 |
0.1732669514 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 15 |
Hb_000853_260 |
0.1744000004 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 7 [Jatropha curcas] |
| 16 |
Hb_013399_080 |
0.1748907089 |
- |
- |
PREDICTED: protein Brevis radix-like 2 [Jatropha curcas] |
| 17 |
Hb_004007_160 |
0.1787272794 |
- |
- |
PREDICTED: formin-like protein 6 [Jatropha curcas] |
| 18 |
Hb_003872_010 |
0.1802684205 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |
| 19 |
Hb_000086_200 |
0.1810059219 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_000322_090 |
0.1829484236 |
- |
- |
diacylglycerol O-acyltransferase 1 [Jatropha curcas] |