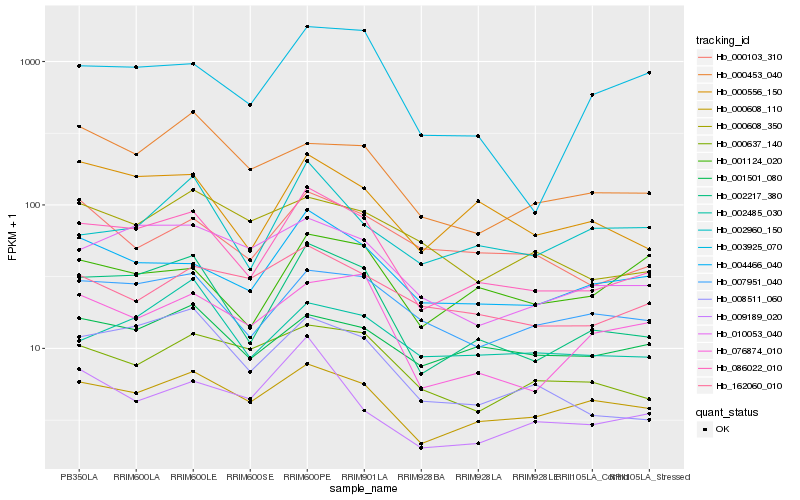
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_086022_010 |
0.0 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 2 |
Hb_002217_380 |
0.058821362 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 3 |
Hb_004466_040 |
0.106508137 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 4 |
Hb_000608_110 |
0.113977866 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 5 |
Hb_000556_150 |
0.1140914133 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 6 |
Hb_008511_060 |
0.1199258269 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 7 |
Hb_007951_040 |
0.1213539228 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 8 |
Hb_010053_040 |
0.1259135065 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 9 |
Hb_076874_010 |
0.1307862633 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 10 |
Hb_000103_310 |
0.1345927 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 11 |
Hb_000637_140 |
0.1388225899 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 12 |
Hb_162060_010 |
0.1411438514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001501_080 |
0.1424690245 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 14 |
Hb_009189_020 |
0.1428373204 |
- |
- |
PREDICTED: CASP-like protein 4B4 [Jatropha curcas] |
| 15 |
Hb_003925_070 |
0.1435024955 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 16 |
Hb_000453_040 |
0.144923323 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 17 |
Hb_002485_030 |
0.1451117401 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 18 |
Hb_002960_150 |
0.1463635984 |
- |
- |
hypothetical protein CISIN_1g029215mg [Citrus sinensis] |
| 19 |
Hb_000608_350 |
0.1468029928 |
- |
- |
PREDICTED: ER membrane protein complex subunit 10 [Jatropha curcas] |
| 20 |
Hb_001124_020 |
0.1472362238 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |