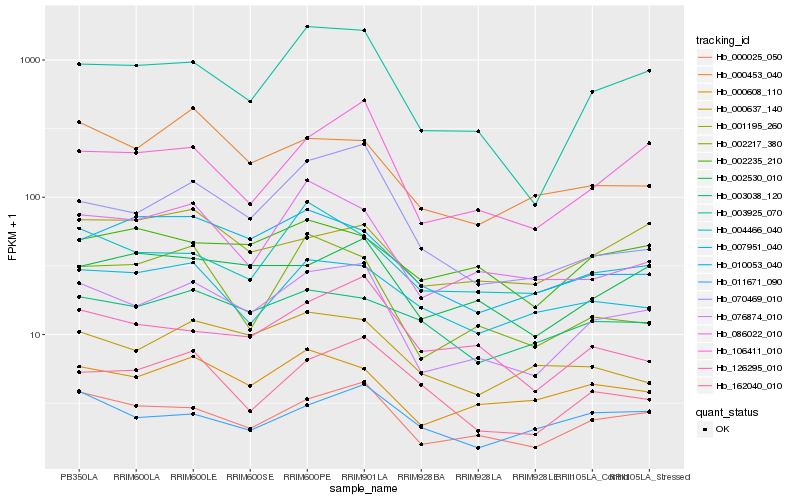
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_076874_010 |
0.0 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 2 |
Hb_000025_050 |
0.0800086485 |
- |
- |
hypothetical protein POPTR_0003s13040g [Populus trichocarpa] |
| 3 |
Hb_003925_070 |
0.0889294352 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 4 |
Hb_106411_010 |
0.1151454546 |
- |
- |
Ribosomal protein S25 family protein [Theobroma cacao] |
| 5 |
Hb_000608_110 |
0.1205566848 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 6 |
Hb_002530_010 |
0.1251441523 |
- |
- |
hypothetical protein EUGRSUZ_C02580 [Eucalyptus grandis] |
| 7 |
Hb_086022_010 |
0.1307862633 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 8 |
Hb_070469_010 |
0.1308748749 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |
| 9 |
Hb_126295_010 |
0.1343887169 |
- |
- |
PREDICTED: protein SEH1 [Jatropha curcas] |
| 10 |
Hb_004466_040 |
0.1360708076 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 11 |
Hb_000637_140 |
0.1377083021 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 12 |
Hb_002217_380 |
0.14005225 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 13 |
Hb_000453_040 |
0.1422489775 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 14 |
Hb_007951_040 |
0.1440730796 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 15 |
Hb_162040_010 |
0.1442146701 |
- |
- |
- |
| 16 |
Hb_010053_040 |
0.1448684499 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 17 |
Hb_011671_090 |
0.1457010535 |
- |
- |
PREDICTED: protein RFT1 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_001195_260 |
0.146370943 |
- |
- |
Mitochondrial 50S ribosomal protein L27 [Hevea brasiliensis] |
| 19 |
Hb_003038_120 |
0.1482704905 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 20 |
Hb_002235_210 |
0.1488436299 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |