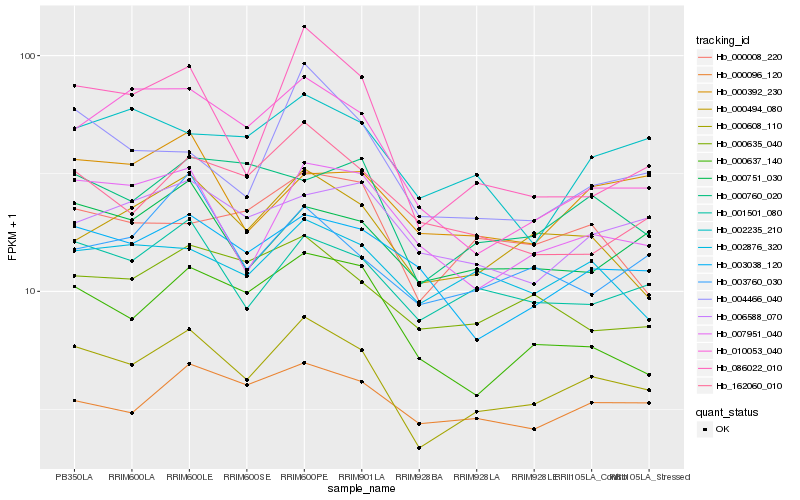
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000608_110 |
0.0 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 2 |
Hb_001501_080 |
0.0912073738 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000751_030 |
0.0993411198 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 4 |
Hb_000494_080 |
0.0996409245 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_007951_040 |
0.1022031157 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 6 |
Hb_000760_020 |
0.1025232881 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF185-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000096_120 |
0.1047090325 |
- |
- |
PREDICTED: uncharacterized protein LOC105639993 [Jatropha curcas] |
| 8 |
Hb_000637_140 |
0.1055530089 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 9 |
Hb_003038_120 |
0.1068876089 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 10 |
Hb_000635_040 |
0.1069841396 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 11 |
Hb_002876_320 |
0.1088995421 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 12 |
Hb_004466_040 |
0.1097121576 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 13 |
Hb_162060_010 |
0.1118835777 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003760_030 |
0.1123411142 |
- |
- |
PREDICTED: novel plant SNARE 13 [Jatropha curcas] |
| 15 |
Hb_000008_220 |
0.1127717964 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 10 [Jatropha curcas] |
| 16 |
Hb_086022_010 |
0.113977866 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 17 |
Hb_006588_070 |
0.1143794977 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 18 |
Hb_002235_210 |
0.1150646093 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |
| 19 |
Hb_010053_040 |
0.1154015232 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 20 |
Hb_000392_230 |
0.1158588484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |