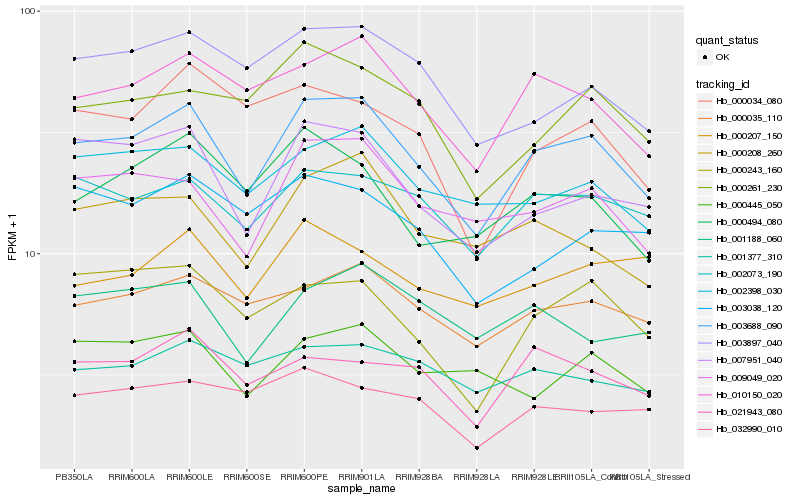
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003688_090 |
0.0 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 2 |
Hb_010150_020 |
0.0767461036 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 3 |
Hb_009049_020 |
0.0812119189 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 4 |
Hb_007951_040 |
0.0826543459 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 5 |
Hb_002073_190 |
0.0842604722 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000035_110 |
0.0862566981 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 7 |
Hb_002398_030 |
0.0875605342 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_000208_260 |
0.0911624417 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 9 |
Hb_003897_040 |
0.0932489408 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 10 |
Hb_000261_230 |
0.0940245456 |
- |
- |
PREDICTED: UBX domain-containing protein 4 [Vitis vinifera] |
| 11 |
Hb_021943_080 |
0.0964241437 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000494_080 |
0.0976085353 |
- |
- |
Palmitoyl-protein thioesterase 1 precursor, putative [Ricinus communis] |
| 13 |
Hb_000243_160 |
0.0984309655 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 14 |
Hb_001377_310 |
0.1020635533 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 15 |
Hb_001188_060 |
0.1022920604 |
- |
- |
- |
| 16 |
Hb_000445_050 |
0.1024718937 |
- |
- |
PREDICTED: DNA topoisomerase 2-binding protein 1 [Jatropha curcas] |
| 17 |
Hb_032990_010 |
0.103623669 |
- |
- |
PREDICTED: fanconi-associated nuclease 1 homolog [Jatropha curcas] |
| 18 |
Hb_000034_080 |
0.1037269817 |
- |
- |
PREDICTED: MADS-box protein AGL24-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_003038_120 |
0.104891147 |
- |
- |
rer1 protein, putative [Ricinus communis] |
| 20 |
Hb_000207_150 |
0.105086765 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |