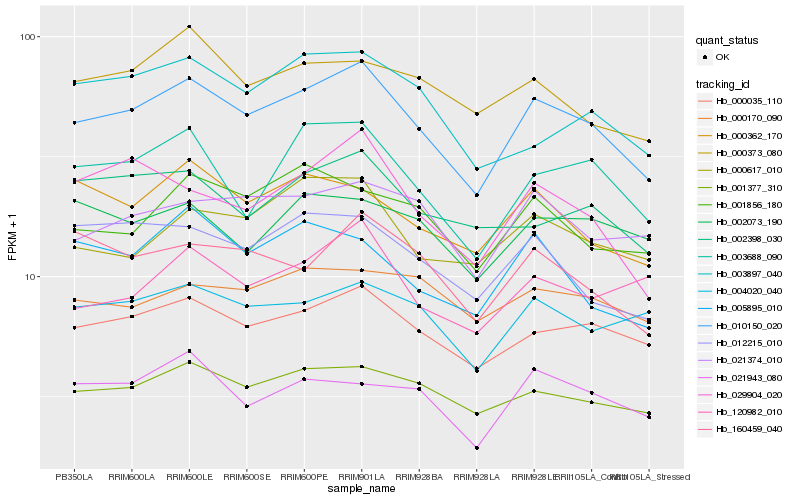
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010150_020 |
0.0 |
transcription factor |
TF Family: HMG |
PREDICTED: FACT complex subunit SSRP1 [Jatropha curcas] |
| 2 |
Hb_000035_110 |
0.06414981 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 3 |
Hb_001377_310 |
0.0729021953 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 4 |
Hb_003688_090 |
0.0767461036 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 5 |
Hb_021374_010 |
0.0827579791 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-A1 [Populus euphratica] |
| 6 |
Hb_029904_020 |
0.0839222014 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 7 |
Hb_004020_040 |
0.0853174495 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000362_170 |
0.0854757533 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 9 |
Hb_160459_040 |
0.0858419471 |
- |
- |
hypothetical protein JCGZ_01511 [Jatropha curcas] |
| 10 |
Hb_001856_180 |
0.0858559347 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 11 |
Hb_000617_010 |
0.0862487252 |
- |
- |
hypothetical protein JCGZ_23296 [Jatropha curcas] |
| 12 |
Hb_021943_080 |
0.0862952966 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase ASHH3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_012215_010 |
0.0864438994 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 14 |
Hb_000373_080 |
0.0877027436 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 15 |
Hb_002398_030 |
0.0882282316 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_005895_010 |
0.0906737709 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003897_040 |
0.0914866363 |
- |
- |
translocon-associated protein, beta subunit precursor, putative [Ricinus communis] |
| 18 |
Hb_120982_010 |
0.0929476051 |
- |
- |
hypothetical protein CICLE_v10003116mg, partial [Citrus clementina] |
| 19 |
Hb_000170_090 |
0.0940259879 |
- |
- |
PREDICTED: UDP-sugar pyrophosphorylase [Jatropha curcas] |
| 20 |
Hb_002073_190 |
0.0945084313 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |