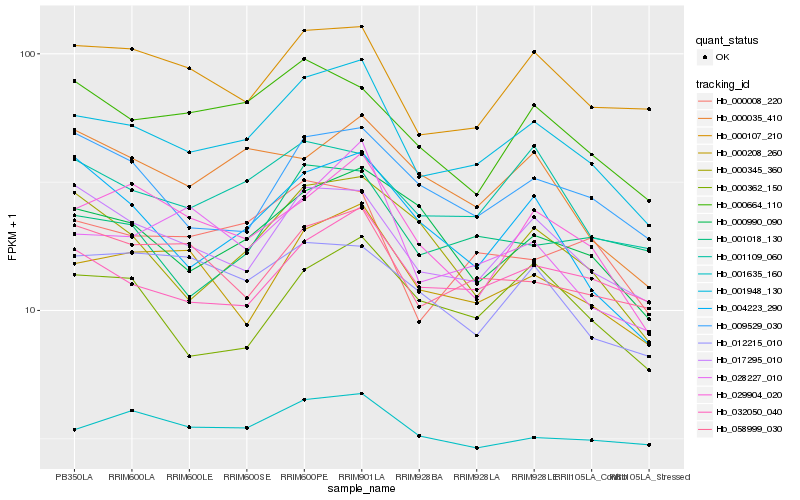
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001948_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 2 |
Hb_017295_010 |
0.076319246 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 3 |
Hb_000208_260 |
0.0816029932 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_001109_060 |
0.0831344322 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_000107_210 |
0.0852733711 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 6 |
Hb_029904_020 |
0.0852994988 |
- |
- |
hypothetical protein JCGZ_06227 [Jatropha curcas] |
| 7 |
Hb_000008_220 |
0.0855150415 |
transcription factor |
TF Family: ARF |
PREDICTED: auxin response factor 10 [Jatropha curcas] |
| 8 |
Hb_032050_040 |
0.0859021238 |
- |
- |
PREDICTED: protein S-acyltransferase 8-like [Jatropha curcas] |
| 9 |
Hb_000990_090 |
0.0893768873 |
- |
- |
PREDICTED: ER membrane protein complex subunit 1 [Jatropha curcas] |
| 10 |
Hb_028227_010 |
0.0905633165 |
- |
- |
PREDICTED: uncharacterized protein LOC105629489 [Jatropha curcas] |
| 11 |
Hb_000664_110 |
0.0905933362 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 12 |
Hb_058999_030 |
0.0914924852 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_009529_030 |
0.0938586687 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 14 |
Hb_000345_360 |
0.0953256533 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 15 |
Hb_000362_150 |
0.0963607742 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 16 |
Hb_012215_010 |
0.0986863167 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 17 |
Hb_004223_290 |
0.0988236 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 18 |
Hb_001635_160 |
0.0995352244 |
- |
- |
hypothetical protein JCGZ_24810 [Jatropha curcas] |
| 19 |
Hb_000035_410 |
0.099657012 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 20 |
Hb_001018_130 |
0.1001329637 |
- |
- |
PREDICTED: uncharacterized protein LOC105638260 [Jatropha curcas] |