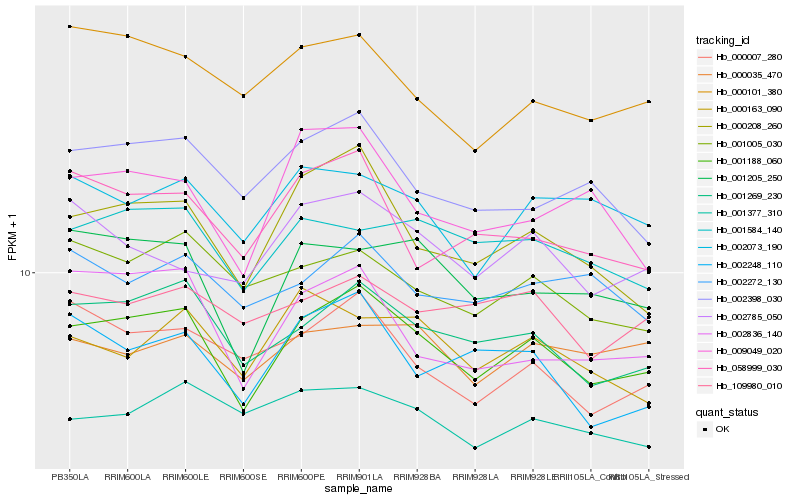
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001188_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_001269_230 |
0.0584174479 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 3 |
Hb_000208_260 |
0.0707716618 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4 [Jatropha curcas] |
| 4 |
Hb_001205_250 |
0.0749531058 |
- |
- |
PREDICTED: uncharacterized protein LOC105643856 [Jatropha curcas] |
| 5 |
Hb_001584_140 |
0.0813784894 |
- |
- |
PREDICTED: uncharacterized protein LOC105647409 [Jatropha curcas] |
| 6 |
Hb_002398_030 |
0.0828662979 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_002248_110 |
0.0853246249 |
- |
- |
hypothetical protein POPTR_0001s41050g [Populus trichocarpa] |
| 8 |
Hb_001005_030 |
0.0857049603 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 9 |
Hb_002836_140 |
0.0860572953 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 10 |
Hb_002073_190 |
0.086107653 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000101_380 |
0.0878244122 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 12 |
Hb_058999_030 |
0.0881427992 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_000007_280 |
0.0891544163 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 14 |
Hb_002785_050 |
0.0901661225 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 15 |
Hb_002272_130 |
0.0911232974 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 16 |
Hb_109980_010 |
0.0911669668 |
- |
- |
PREDICTED: uncharacterized protein LOC105647182 isoform X1 [Jatropha curcas] |
| 17 |
Hb_009049_020 |
0.0912398514 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 18 |
Hb_000163_090 |
0.0913979259 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 19 |
Hb_001377_310 |
0.0916239234 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 20 |
Hb_000035_470 |
0.0925976856 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |