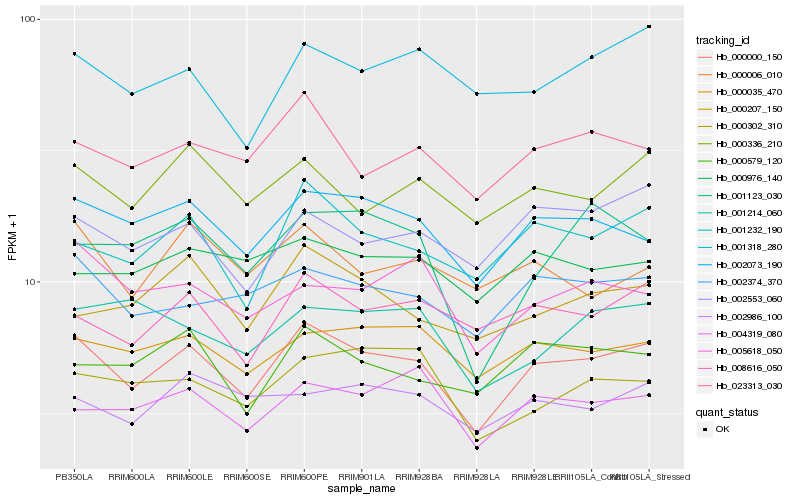
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000000_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 2 |
Hb_002073_190 |
0.0709390505 |
- |
- |
PREDICTED: uncharacterized protein LOC105649812 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001232_190 |
0.0769373659 |
- |
- |
PREDICTED: uncharacterized protein LOC105639761 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002374_370 |
0.0815946608 |
- |
- |
protein with unknown function [Ricinus communis] |
| 5 |
Hb_023313_030 |
0.0844437917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000035_470 |
0.0856644807 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 7 |
Hb_008616_050 |
0.0859865054 |
- |
- |
PREDICTED: putative deoxyribonuclease TATDN1 [Jatropha curcas] |
| 8 |
Hb_000336_210 |
0.0871983347 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 9 |
Hb_002553_060 |
0.0885554972 |
- |
- |
PREDICTED: electron transfer flavoprotein subunit beta, mitochondrial [Jatropha curcas] |
| 10 |
Hb_004319_080 |
0.0896767127 |
- |
- |
PREDICTED: GPI ethanolamine phosphate transferase 1 [Jatropha curcas] |
| 11 |
Hb_000579_120 |
0.0909907298 |
- |
- |
PREDICTED: uncharacterized protein LOC105633845 [Jatropha curcas] |
| 12 |
Hb_001318_280 |
0.0932047192 |
- |
- |
Protein FAM18B, putative [Ricinus communis] |
| 13 |
Hb_000207_150 |
0.0946212752 |
- |
- |
PREDICTED: uncharacterized membrane protein At4g09580 [Jatropha curcas] |
| 14 |
Hb_005618_050 |
0.0949795368 |
- |
- |
PREDICTED: charged multivesicular body protein 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000302_310 |
0.0954938962 |
- |
- |
PREDICTED: glutaminyl-peptide cyclotransferase-like [Jatropha curcas] |
| 16 |
Hb_002986_100 |
0.0956321735 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 17 |
Hb_000006_010 |
0.096899711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000976_140 |
0.0986500964 |
- |
- |
PREDICTED: uncharacterized protein LOC105646208 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001123_030 |
0.0997425394 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_001214_060 |
0.0998297607 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC8 [Jatropha curcas] |