| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
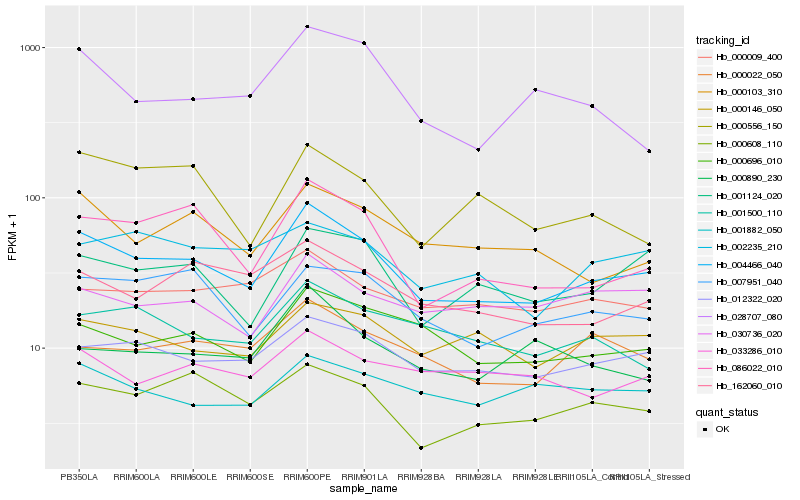
Hb_004466_040 |
0.0 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 2 |
Hb_012322_020 |
0.1046567043 |
- |
- |
- |
| 3 |
Hb_086022_010 |
0.106508137 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 4 |
Hb_000103_310 |
0.1089047924 |
- |
- |
PREDICTED: transmembrane emp24 domain-containing protein p24beta3 [Jatropha curcas] |
| 5 |
Hb_000608_110 |
0.1097121576 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 6 |
Hb_000696_010 |
0.1098710563 |
- |
- |
RING-H2 finger protein ATL4M, putative [Ricinus communis] |
| 7 |
Hb_001124_020 |
0.1118396342 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Jatropha curcas] |
| 8 |
Hb_007951_040 |
0.1134622503 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 9 |
Hb_162060_010 |
0.1165461901 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000022_050 |
0.118421454 |
- |
- |
PREDICTED: retinol dehydrogenase 11-like isoform X2 [Glycine max] |
| 11 |
Hb_000146_050 |
0.1223069828 |
- |
- |
PREDICTED: protein unc-50 homolog isoform X2 [Gossypium raimondii] |
| 12 |
Hb_001500_110 |
0.1231019402 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_002235_210 |
0.1276228508 |
- |
- |
PREDICTED: vesicle transport v-SNARE 13-like [Populus euphratica] |
| 14 |
Hb_000009_400 |
0.128289348 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 15 |
Hb_030736_020 |
0.1290346155 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_033286_010 |
0.1290540514 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 14 [Jatropha curcas] |
| 17 |
Hb_028707_080 |
0.1293283244 |
- |
- |
plasma membrane aquaporin 1 [Hevea brasiliensis] |
| 18 |
Hb_001882_050 |
0.1300211257 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_000890_230 |
0.1300572988 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 20 |
Hb_000556_150 |
0.1301444925 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |