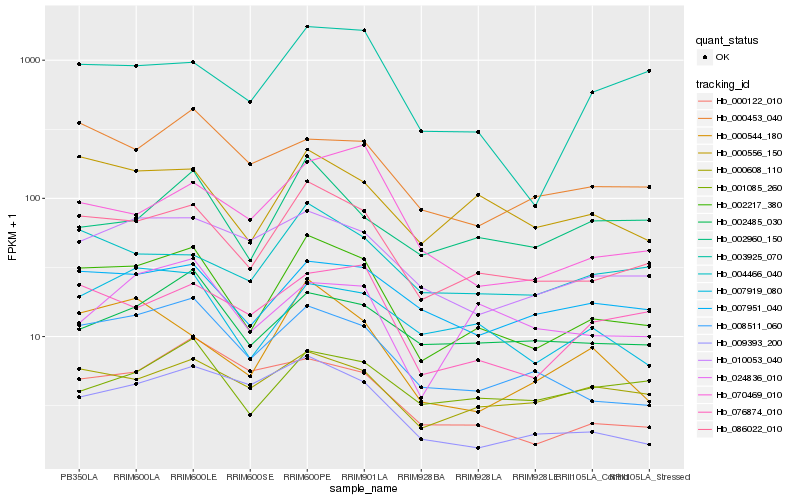
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_380 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643567 [Jatropha curcas] |
| 2 |
Hb_086022_010 |
0.058821362 |
- |
- |
PREDICTED: succinate dehydrogenase assembly factor 1 homolog, mitochondrial [Nicotiana tomentosiformis] |
| 3 |
Hb_008511_060 |
0.1190361977 |
- |
- |
PREDICTED: probable receptor-like serine/threonine-protein kinase At4g34500 [Jatropha curcas] |
| 4 |
Hb_000556_150 |
0.126318871 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 5 |
Hb_000608_110 |
0.139010553 |
- |
- |
PREDICTED: probable tyrosine-protein phosphatase At1g05000 [Jatropha curcas] |
| 6 |
Hb_007919_080 |
0.1397726984 |
- |
- |
PREDICTED: uncharacterized protein At1g76660 isoform X1 [Jatropha curcas] |
| 7 |
Hb_076874_010 |
0.14005225 |
- |
- |
PREDICTED: ER lumen protein-retaining receptor [Jatropha curcas] |
| 8 |
Hb_007951_040 |
0.1405899914 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 9 |
Hb_010053_040 |
0.1420569041 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 10 |
Hb_004466_040 |
0.1478242277 |
- |
- |
PREDICTED: protein RER1B-like [Gossypium raimondii] |
| 11 |
Hb_003925_070 |
0.1496466051 |
- |
- |
ATOZI1, putative [Ricinus communis] |
| 12 |
Hb_000453_040 |
0.1511971128 |
- |
- |
Triosephosphate isomerase, cytosolic [Gossypium arboreum] |
| 13 |
Hb_000544_180 |
0.1521690754 |
- |
- |
PREDICTED: probable auxin efflux carrier component 1c isoform X1 [Jatropha curcas] |
| 14 |
Hb_002485_030 |
0.1538779852 |
- |
- |
PREDICTED: 2-dehydro-3-deoxyphosphooctonate aldolase 1-like [Jatropha curcas] |
| 15 |
Hb_009393_200 |
0.1546986254 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 16 |
Hb_001085_260 |
0.1575861137 |
- |
- |
PREDICTED: uncharacterized protein LOC105633184 [Jatropha curcas] |
| 17 |
Hb_002960_150 |
0.1622599814 |
- |
- |
hypothetical protein CISIN_1g029215mg [Citrus sinensis] |
| 18 |
Hb_000122_010 |
0.1650399438 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 19 |
Hb_024836_010 |
0.1654830269 |
- |
- |
Rho GDP-dissociation inhibitor, putative [Ricinus communis] |
| 20 |
Hb_070469_010 |
0.1656237789 |
- |
- |
hypothetical protein POPTR_0002s21850g [Populus trichocarpa] |