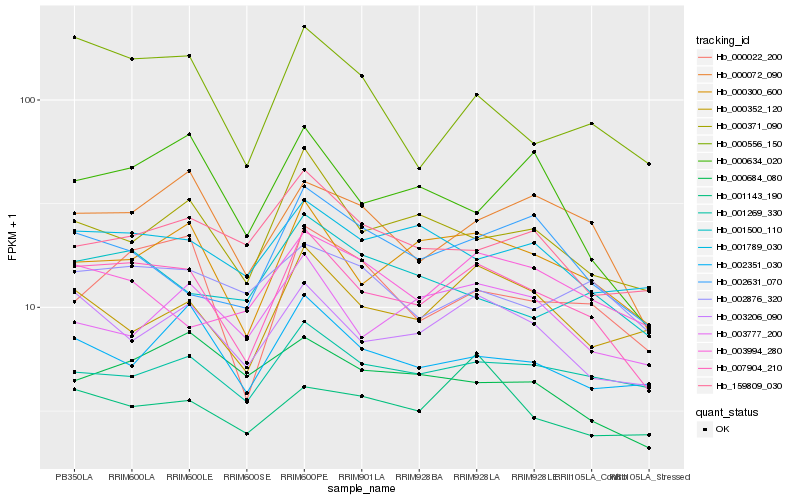
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007904_210 |
0.0 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 1D [Jatropha curcas] |
| 2 |
Hb_000300_600 |
0.0988283279 |
- |
- |
tubulin alpha chain, putative [Ricinus communis] |
| 3 |
Hb_003206_090 |
0.1019979718 |
- |
- |
PREDICTED: protein trichome birefringence-like 5 [Jatropha curcas] |
| 4 |
Hb_000072_090 |
0.115587703 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 5 |
Hb_000371_090 |
0.1197693313 |
- |
- |
PREDICTED: dolichyl-diphosphooligosaccharide--protein glycosyltransferase 48 kDa subunit-like [Jatropha curcas] |
| 6 |
Hb_000022_200 |
0.1236633576 |
- |
- |
PREDICTED: uncharacterized protein LOC105638292 [Jatropha curcas] |
| 7 |
Hb_000556_150 |
0.1244985579 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 8 |
Hb_000352_120 |
0.1283773089 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105630380 [Jatropha curcas] |
| 9 |
Hb_002351_030 |
0.1297917223 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_001789_030 |
0.1301544045 |
- |
- |
PREDICTED: signal peptide peptidase-like 1 [Jatropha curcas] |
| 11 |
Hb_001269_330 |
0.1303564922 |
- |
- |
PREDICTED: uncharacterized protein LOC105645377 [Jatropha curcas] |
| 12 |
Hb_002631_070 |
0.1310194203 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 13 |
Hb_000634_020 |
0.1334802241 |
- |
- |
hypothetical protein JCGZ_11888 [Jatropha curcas] |
| 14 |
Hb_003777_200 |
0.1369794528 |
- |
- |
PREDICTED: uncharacterized protein LOC105640933 [Jatropha curcas] |
| 15 |
Hb_002876_320 |
0.1377015222 |
- |
- |
PREDICTED: uncharacterized protein LOC105632052 [Jatropha curcas] |
| 16 |
Hb_001500_110 |
0.1388220916 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_159809_030 |
0.1397166894 |
- |
- |
PREDICTED: UDP-D-xylose:L-fucose alpha-1,3-D-xylosyltransferase 2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_003994_280 |
0.1405629345 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 19 |
Hb_000684_080 |
0.1406932197 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase VIII.1 [Jatropha curcas] |
| 20 |
Hb_001143_190 |
0.1412361248 |
- |
- |
PREDICTED: uncharacterized protein LOC105638976 [Jatropha curcas] |