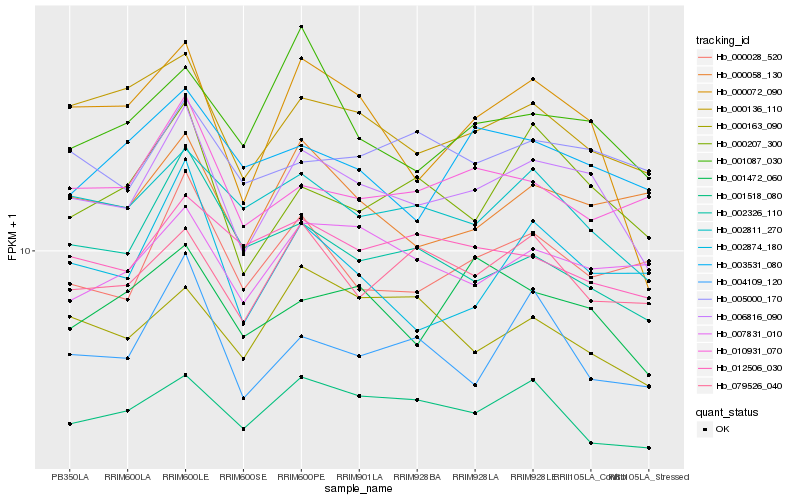
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_090 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647425 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000136_110 |
0.0828306501 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 3 |
Hb_002811_270 |
0.0841022038 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 4 |
Hb_000072_090 |
0.0847333382 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 5 |
Hb_000207_300 |
0.0855329145 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_001472_060 |
0.0922261945 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000028_520 |
0.0926961184 |
- |
- |
hypothetical protein L484_025125 [Morus notabilis] |
| 8 |
Hb_001518_080 |
0.0928741932 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 9 |
Hb_079526_040 |
0.0943158096 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 10 |
Hb_004109_120 |
0.0957761825 |
- |
- |
PREDICTED: kinesin-related protein 6 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000058_130 |
0.0962220353 |
- |
- |
PREDICTED: uncharacterized protein LOC105640884 [Jatropha curcas] |
| 12 |
Hb_007831_010 |
0.0967898105 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 13 |
Hb_010931_070 |
0.0972873558 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002874_180 |
0.0978159373 |
- |
- |
PREDICTED: protein AIG2-like isoform X1 [Jatropha curcas] |
| 15 |
Hb_005000_170 |
0.0981096201 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 16 |
Hb_001087_030 |
0.0996134872 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP2A catalytic subunit [Jatropha curcas] |
| 17 |
Hb_012506_030 |
0.1005522927 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 18 |
Hb_002326_110 |
0.1011167892 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 19 |
Hb_003531_080 |
0.1016866098 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000163_090 |
0.1030767362 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |