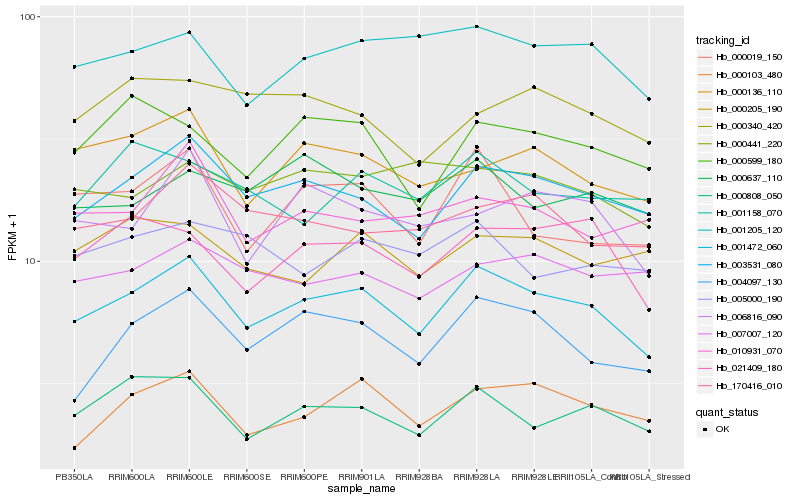
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001472_060 |
0.0 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 2 |
Hb_003531_080 |
0.0638531664 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 3 |
Hb_004097_130 |
0.084045369 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 4 |
Hb_021409_180 |
0.0844987924 |
- |
- |
PREDICTED: glutamine-dependent NAD(+) synthetase [Jatropha curcas] |
| 5 |
Hb_000808_050 |
0.086151246 |
- |
- |
exonuclease, putative [Ricinus communis] |
| 6 |
Hb_001205_120 |
0.088339211 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 7 |
Hb_000019_150 |
0.0885426224 |
- |
- |
PREDICTED: uncharacterized protein LOC105630208 [Jatropha curcas] |
| 8 |
Hb_000136_110 |
0.0904595099 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 9 |
Hb_000103_480 |
0.091685051 |
- |
- |
- |
| 10 |
Hb_006816_090 |
0.0922261945 |
- |
- |
PREDICTED: uncharacterized protein LOC105647425 isoform X2 [Jatropha curcas] |
| 11 |
Hb_170416_010 |
0.092777692 |
- |
- |
PREDICTED: cold-regulated 413 plasma membrane protein 2-like [Jatropha curcas] |
| 12 |
Hb_000599_180 |
0.0956000867 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 13 |
Hb_010931_070 |
0.0992078259 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007007_120 |
0.1000090868 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000637_110 |
0.1014328891 |
- |
- |
hypothetical protein B456_013G081300 [Gossypium raimondii] |
| 16 |
Hb_000441_220 |
0.102528904 |
- |
- |
PREDICTED: histidinol dehydrogenase, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000340_420 |
0.1038479184 |
- |
- |
PREDICTED: LEC14B protein isoform X2 [Jatropha curcas] |
| 18 |
Hb_001158_070 |
0.1042350219 |
- |
- |
Protein DEK, putative [Ricinus communis] |
| 19 |
Hb_000205_190 |
0.1050026864 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 20 |
Hb_005000_190 |
0.1056763658 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 64 [Jatropha curcas] |