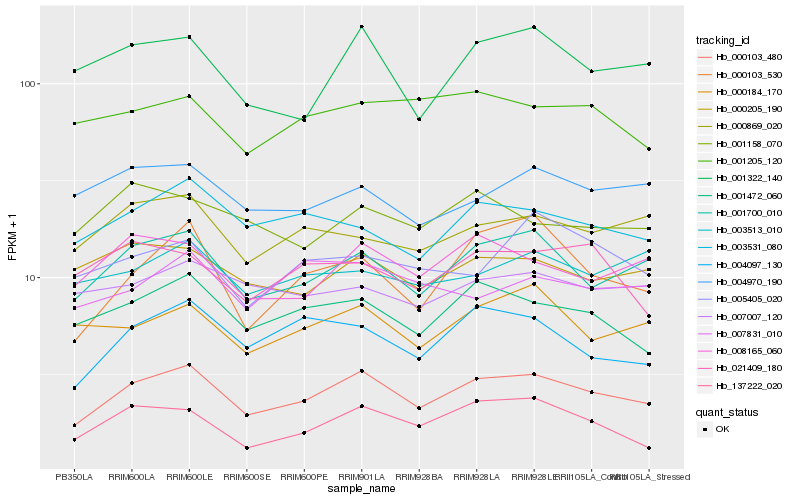
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_480 |
0.0 |
- |
- |
- |
| 2 |
Hb_001700_010 |
0.0710721671 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 3 |
Hb_001472_060 |
0.091685051 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 4 |
Hb_004097_130 |
0.09291542 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 5 |
Hb_000103_530 |
0.0952702281 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 6 |
Hb_008165_060 |
0.1000327237 |
- |
- |
PREDICTED: uncharacterized protein LOC105635541 [Jatropha curcas] |
| 7 |
Hb_001322_140 |
0.102029476 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_137222_020 |
0.1040008092 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 9 |
Hb_003531_080 |
0.1042635213 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000184_170 |
0.1066357182 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 11 |
Hb_000869_020 |
0.1090370714 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 12 |
Hb_000205_190 |
0.1105631721 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_004970_190 |
0.1148803085 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_001205_120 |
0.1164814534 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 15 |
Hb_007007_120 |
0.1175533865 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003513_010 |
0.1178696287 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 17 |
Hb_007831_010 |
0.1180807779 |
- |
- |
PREDICTED: oligoribonuclease [Vitis vinifera] |
| 18 |
Hb_021409_180 |
0.1188130339 |
- |
- |
PREDICTED: glutamine-dependent NAD(+) synthetase [Jatropha curcas] |
| 19 |
Hb_005405_020 |
0.1194606679 |
- |
- |
PREDICTED: petal death protein isoform X1 [Jatropha curcas] |
| 20 |
Hb_001158_070 |
0.1195849255 |
- |
- |
Protein DEK, putative [Ricinus communis] |