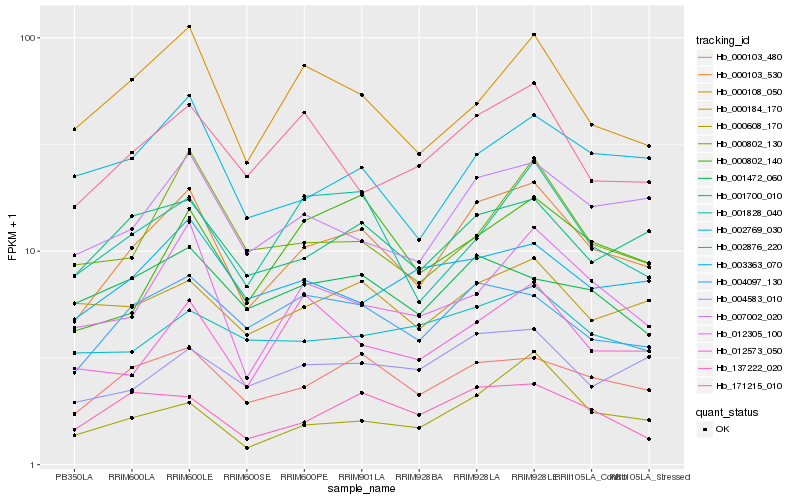
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_530 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 2 |
Hb_000103_480 |
0.0952702281 |
- |
- |
- |
| 3 |
Hb_007002_020 |
0.1023862916 |
- |
- |
PREDICTED: uncharacterized protein LOC105642604 [Jatropha curcas] |
| 4 |
Hb_001700_010 |
0.1083783663 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 5 |
Hb_004097_130 |
0.1150795029 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 6 |
Hb_012305_100 |
0.1193188892 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_004583_010 |
0.1210345942 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g22150, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000108_050 |
0.1228757843 |
- |
- |
PREDICTED: mitochondrial dicarboxylate/tricarboxylate transporter DTC [Jatropha curcas] |
| 9 |
Hb_137222_020 |
0.1251258508 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 10 |
Hb_171215_010 |
0.1263805192 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 11 |
Hb_002876_220 |
0.1277737862 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |
| 12 |
Hb_001828_040 |
0.1293922858 |
- |
- |
PREDICTED: F-box protein SKIP14 [Jatropha curcas] |
| 13 |
Hb_000802_130 |
0.1311064402 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 14 |
Hb_012573_050 |
0.1313932374 |
- |
- |
choline/ethanolamine kinase family protein [Populus trichocarpa] |
| 15 |
Hb_000184_170 |
0.1313987156 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 16 |
Hb_003363_070 |
0.1322559629 |
- |
- |
- |
| 17 |
Hb_000802_140 |
0.1325305011 |
- |
- |
hypothetical protein B456_011G045700 [Gossypium raimondii] |
| 18 |
Hb_000608_170 |
0.132592986 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 19 |
Hb_002769_030 |
0.1365468695 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_001472_060 |
0.1372707535 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |