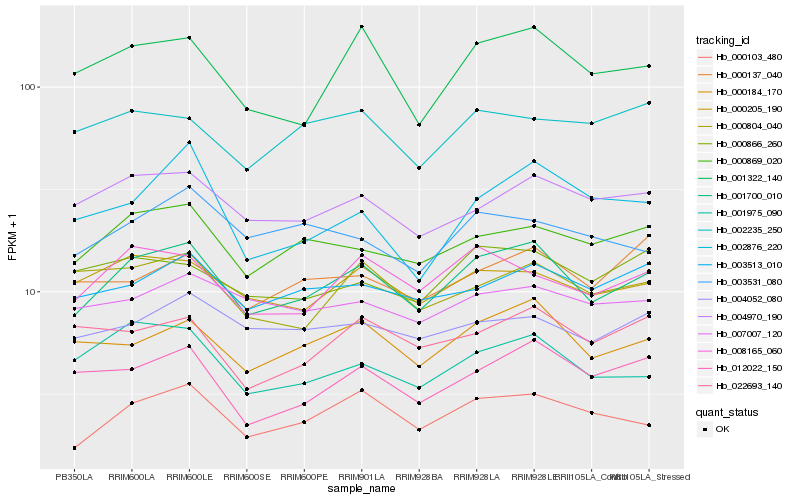
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001700_010 |
0.0 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 2 |
Hb_000103_480 |
0.0710721671 |
- |
- |
- |
| 3 |
Hb_000184_170 |
0.0750279883 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 4 |
Hb_000869_020 |
0.0810817082 |
- |
- |
PREDICTED: uncharacterized protein LOC105641036 [Jatropha curcas] |
| 5 |
Hb_008165_060 |
0.0813618014 |
- |
- |
PREDICTED: uncharacterized protein LOC105635541 [Jatropha curcas] |
| 6 |
Hb_001322_140 |
0.0818843223 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000205_190 |
0.085714136 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_004970_190 |
0.0860696652 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_001975_090 |
0.0869669419 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 10 |
Hb_003513_010 |
0.0895252341 |
- |
- |
Structural maintenance of chromosome 1 protein, putative [Ricinus communis] |
| 11 |
Hb_012022_150 |
0.09039151 |
- |
- |
PREDICTED: protein root UVB sensitive 5 [Jatropha curcas] |
| 12 |
Hb_004052_080 |
0.0952936611 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000866_260 |
0.0967087551 |
- |
- |
PREDICTED: F-box/kelch-repeat protein SKIP4 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000804_040 |
0.0971316113 |
- |
- |
protein with unknown function [Ricinus communis] |
| 15 |
Hb_007007_120 |
0.0985806852 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 16 |
Hb_003531_080 |
0.1003803089 |
- |
- |
PREDICTED: small G protein signaling modulator 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_022693_140 |
0.1011212946 |
- |
- |
PREDICTED: uncharacterized protein LOC105632654 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000137_040 |
0.1014130872 |
- |
- |
aspartyl-tRNA synthetase, putative [Ricinus communis] |
| 19 |
Hb_002235_250 |
0.1040456761 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002876_220 |
0.1041960578 |
- |
- |
PREDICTED: uncharacterized protein LOC105633932 [Jatropha curcas] |