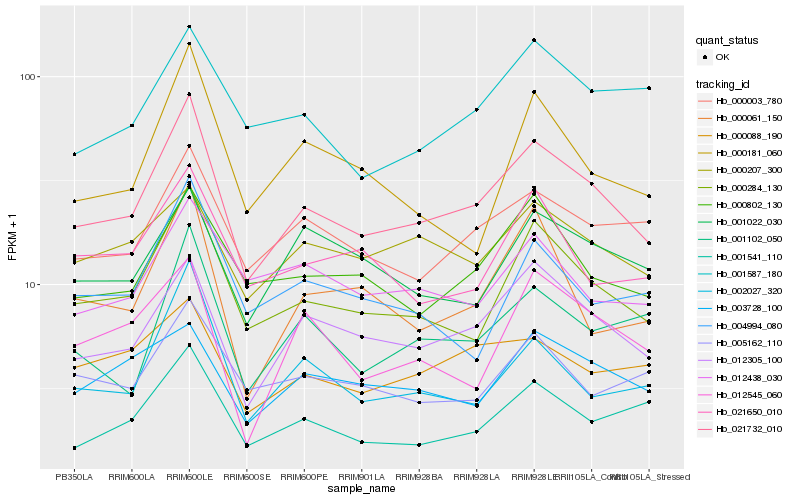
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021732_010 |
0.0 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 2 |
Hb_000284_130 |
0.0811396703 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 3 |
Hb_012305_100 |
0.0852015235 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000003_780 |
0.1094743621 |
- |
- |
hexokinase [Manihot esculenta] |
| 5 |
Hb_000802_130 |
0.1122766109 |
- |
- |
hypothetical protein JCGZ_04636 [Jatropha curcas] |
| 6 |
Hb_001541_110 |
0.118342266 |
- |
- |
PREDICTED: sufE-like protein 2, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_002027_320 |
0.1197050372 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 8 |
Hb_001102_050 |
0.1208468455 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 9 |
Hb_000061_150 |
0.1240558505 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003728_100 |
0.1247419527 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000207_300 |
0.124951446 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_021650_010 |
0.1256289469 |
- |
- |
hypothetical protein JCGZ_09648 [Jatropha curcas] |
| 13 |
Hb_000088_190 |
0.1262087919 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001022_030 |
0.1280088223 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 15 |
Hb_000181_060 |
0.1291520451 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005162_110 |
0.1293497226 |
- |
- |
PREDICTED: uncharacterized protein LOC105648430 [Jatropha curcas] |
| 17 |
Hb_001587_180 |
0.1301458868 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase B5-like [Jatropha curcas] |
| 18 |
Hb_004994_080 |
0.1304162459 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_012438_030 |
0.1314031492 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 20 |
Hb_012545_060 |
0.1314636826 |
- |
- |
PREDICTED: uncharacterized protein LOC103986927 [Musa acuminata subsp. malaccensis] |