| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
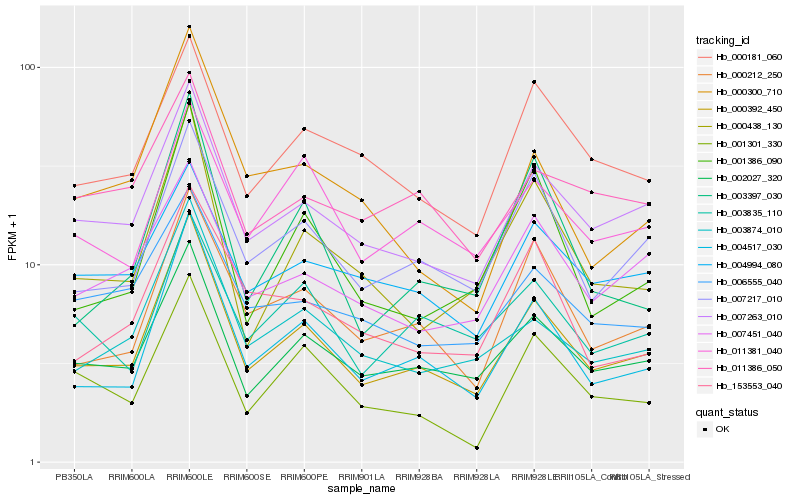
Hb_000392_450 |
0.0 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004517_030 |
0.0617887328 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_007263_010 |
0.0801205096 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 4 |
Hb_002027_320 |
0.0805999333 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 5 |
Hb_003874_010 |
0.0945044041 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 6 |
Hb_007217_010 |
0.0961886492 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000212_250 |
0.1007628533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000438_130 |
0.1108782703 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 9 |
Hb_001386_090 |
0.114708194 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 10 |
Hb_001301_330 |
0.1207303937 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 11 |
Hb_007451_040 |
0.1246007668 |
- |
- |
PREDICTED: uncharacterized protein LOC105643411 [Jatropha curcas] |
| 12 |
Hb_000181_060 |
0.1246991305 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 13 |
Hb_011381_040 |
0.1266375329 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003397_030 |
0.1279488441 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_004994_080 |
0.1287205414 |
- |
- |
PREDICTED: cell division protein FtsY homolog, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_006555_040 |
0.1289672004 |
- |
- |
PREDICTED: uncharacterized protein LOC105639405 [Jatropha curcas] |
| 17 |
Hb_003835_110 |
0.1295942125 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 18 |
Hb_153553_040 |
0.1306008003 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 19 |
Hb_000300_710 |
0.1315033579 |
- |
- |
PREDICTED: uncharacterized protein LOC105123549 [Populus euphratica] |
| 20 |
Hb_011386_050 |
0.1352214445 |
- |
- |
PREDICTED: malonyl-CoA-acyl carrier protein transacylase, mitochondrial [Jatropha curcas] |