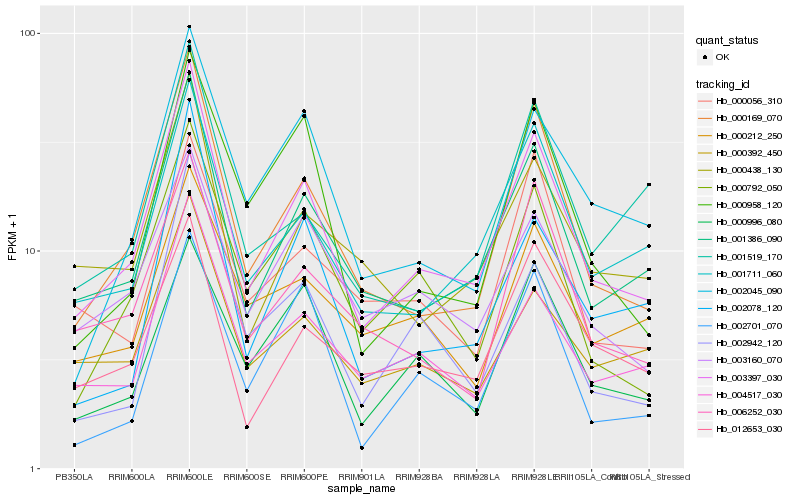
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003397_030 |
0.0 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 2 |
Hb_001386_090 |
0.0764776702 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 3 |
Hb_000169_070 |
0.081803629 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 4 |
Hb_001711_060 |
0.1105948908 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 5 |
Hb_004517_030 |
0.1107583122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000792_050 |
0.1245388369 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_000438_130 |
0.1255596925 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 8 |
Hb_003160_070 |
0.127460372 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 9 |
Hb_000392_450 |
0.1279488441 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_006252_030 |
0.1289029577 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 11 |
Hb_002045_090 |
0.1290105711 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 12 |
Hb_012653_030 |
0.1351314599 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000996_080 |
0.1369628226 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 14 |
Hb_002078_120 |
0.138025391 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 15 |
Hb_000212_250 |
0.1406695227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002942_120 |
0.1417468648 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 17 |
Hb_000958_120 |
0.1456317166 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 18 |
Hb_001519_170 |
0.1463651292 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002701_070 |
0.1465473756 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 20 |
Hb_000056_310 |
0.1466350382 |
- |
- |
PREDICTED: uncharacterized protein LOC105630433 [Jatropha curcas] |