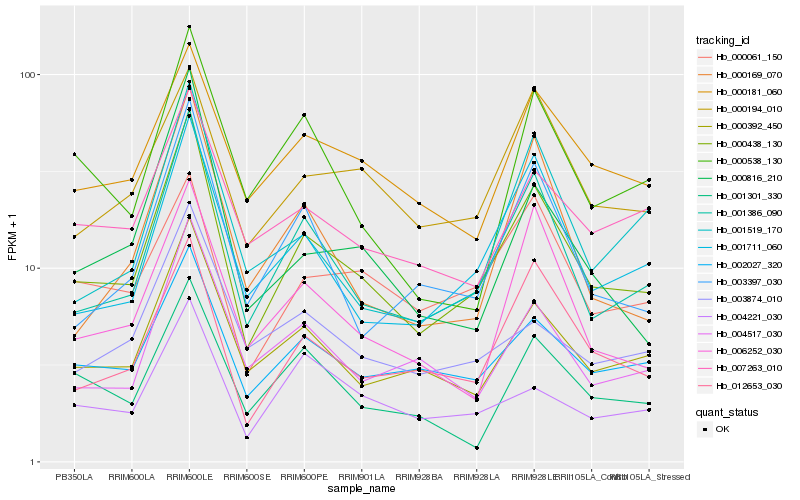
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000438_130 |
0.0 |
- |
- |
uroporphyrinogen decarboxylase, putative [Ricinus communis] |
| 2 |
Hb_001386_090 |
0.0779877746 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 3 |
Hb_000392_450 |
0.1108782703 |
- |
- |
PREDICTED: isochorismate synthase 2, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001711_060 |
0.1194126823 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 5 |
Hb_002027_320 |
0.1207533535 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 6 |
Hb_012653_030 |
0.1237918856 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_003397_030 |
0.1255596925 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004517_030 |
0.1280091487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_007263_010 |
0.1382959577 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 10 |
Hb_000181_060 |
0.1387245012 |
- |
- |
PREDICTED: 30S ribosomal protein S20, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000816_210 |
0.1395205587 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase isoform X2 [Jatropha curcas] |
| 12 |
Hb_006252_030 |
0.1397599894 |
- |
- |
PREDICTED: uncharacterized protein LOC105638333 [Jatropha curcas] |
| 13 |
Hb_000169_070 |
0.1420377973 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 14 |
Hb_000194_010 |
0.1425447136 |
- |
- |
omega-6 fatty acid desaturase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001519_170 |
0.1438325912 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001301_330 |
0.1439104557 |
- |
- |
OTU-like cysteine protease family protein [Populus trichocarpa] |
| 17 |
Hb_003874_010 |
0.1439696729 |
transcription factor |
TF Family: B3 |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_004221_030 |
0.1488906285 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000538_130 |
0.149030404 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 20 |
Hb_000061_150 |
0.1500441843 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |