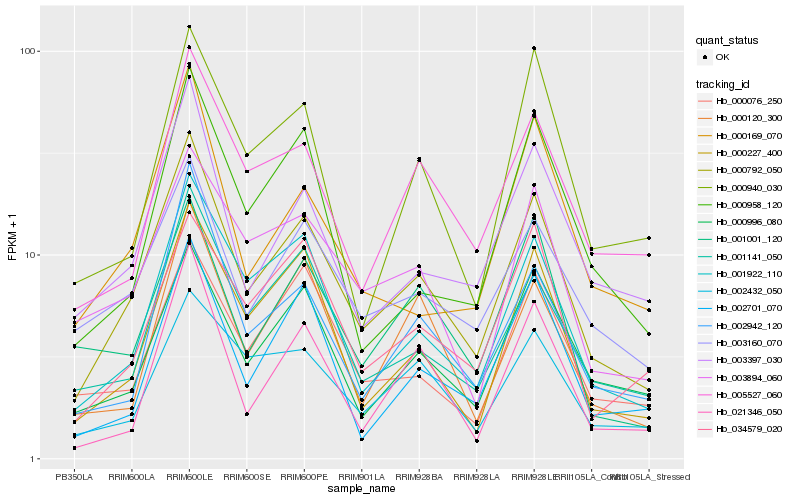
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000792_050 |
0.0 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_001922_110 |
0.107712771 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000227_400 |
0.1192022539 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 4 |
Hb_001141_050 |
0.120139463 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 5 |
Hb_003160_070 |
0.1213330747 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |
| 6 |
Hb_003397_030 |
0.1245388369 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000996_080 |
0.1269768123 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 8 |
Hb_005527_060 |
0.128382684 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 9 |
Hb_002942_120 |
0.1330873278 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 10 |
Hb_003894_060 |
0.13313423 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002701_070 |
0.1369085328 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 12 |
Hb_000120_300 |
0.1401486277 |
- |
- |
PREDICTED: kinesin-like calmodulin-binding protein isoform X1 [Eucalyptus grandis] |
| 13 |
Hb_002432_050 |
0.140153956 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 14 |
Hb_021346_050 |
0.1412930804 |
- |
- |
xylulose kinase, putative [Ricinus communis] |
| 15 |
Hb_000958_120 |
0.1418603334 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 16 |
Hb_000076_250 |
0.1419087126 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000169_070 |
0.1427441922 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 18 |
Hb_034579_020 |
0.1443726306 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001001_120 |
0.1443864201 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 20 |
Hb_000940_030 |
0.145822889 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |