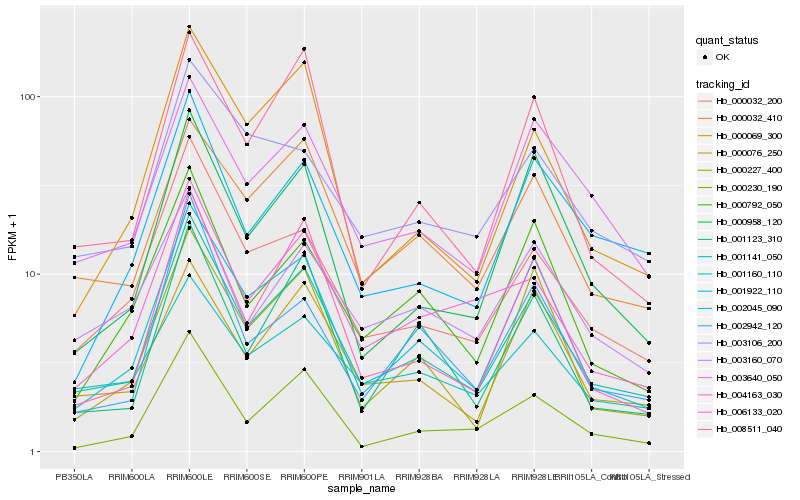
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001141_050 |
0.0 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_001922_110 |
0.0823777101 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000032_410 |
0.1031862977 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 4 |
Hb_000032_200 |
0.1044208842 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 5 |
Hb_006133_020 |
0.1112500753 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 6 |
Hb_000076_250 |
0.1152385444 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003640_050 |
0.1162554207 |
- |
- |
PREDICTED: protein LUTEIN DEFICIENT 5, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000230_190 |
0.1172135184 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000227_400 |
0.1178585602 |
- |
- |
lipoxygenase, putative [Ricinus communis] |
| 10 |
Hb_008511_040 |
0.1189180685 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 11 |
Hb_004163_030 |
0.1200274548 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 12 |
Hb_000792_050 |
0.120139463 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic isoform X1 [Jatropha curcas] |
| 13 |
Hb_000958_120 |
0.1204868631 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 14 |
Hb_000069_300 |
0.1210163694 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_001160_110 |
0.1244553146 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 16 |
Hb_001123_310 |
0.1249651 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 17 |
Hb_002045_090 |
0.129600025 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 18 |
Hb_003106_200 |
0.132822459 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002942_120 |
0.1330832273 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 20 |
Hb_003160_070 |
0.1341740868 |
- |
- |
PREDICTED: VIN3-like protein 1 [Jatropha curcas] |