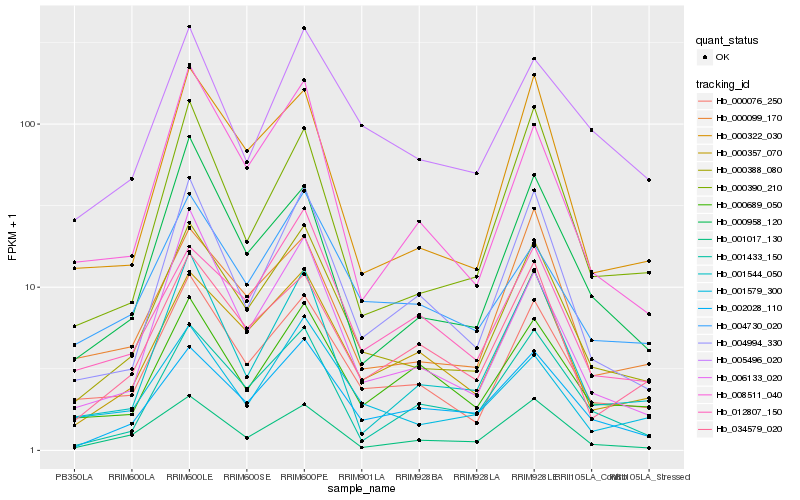
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000388_080 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 2 |
Hb_000076_250 |
0.0904381277 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000357_070 |
0.0991747558 |
- |
- |
- |
| 4 |
Hb_002028_110 |
0.1078962889 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 5 |
Hb_000322_030 |
0.1093134761 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 6 |
Hb_004994_330 |
0.1146061641 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_001579_300 |
0.1194735845 |
- |
- |
PREDICTED: probable polygalacturonase At1g80170 [Jatropha curcas] |
| 8 |
Hb_034579_020 |
0.120254365 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000099_170 |
0.1226294565 |
- |
- |
PREDICTED: uncharacterized protein LOC105645733 [Jatropha curcas] |
| 10 |
Hb_004730_020 |
0.1244565088 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 11 |
Hb_001017_130 |
0.1284797127 |
- |
- |
hypothetical protein POPTR_0010s06070g [Populus trichocarpa] |
| 12 |
Hb_001433_150 |
0.1285582673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_006133_020 |
0.1301145651 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 [Jatropha curcas] |
| 14 |
Hb_000390_210 |
0.1304258883 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 15 |
Hb_000958_120 |
0.1305317507 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 16 |
Hb_005496_020 |
0.1315943987 |
- |
- |
PREDICTED: uncharacterized protein LOC105630867 [Jatropha curcas] |
| 17 |
Hb_008511_040 |
0.1316267088 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 18 |
Hb_012807_150 |
0.1325400346 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 19 |
Hb_000689_050 |
0.1326078432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001544_050 |
0.1328420509 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |