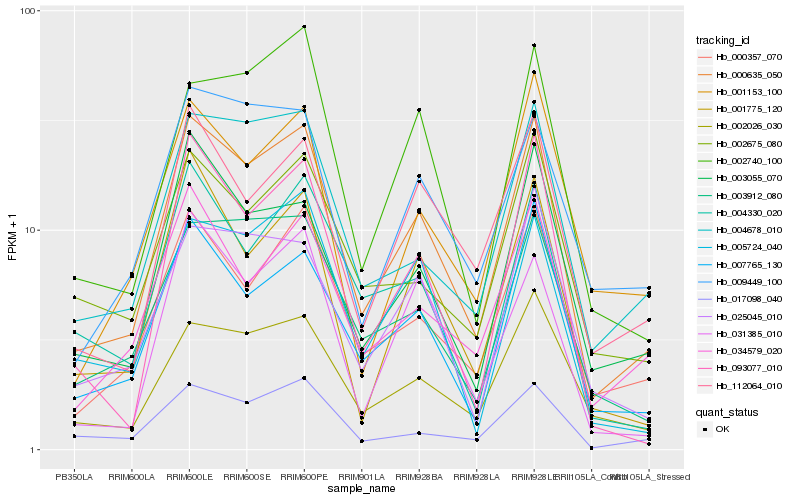
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000635_050 |
0.0 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 2 |
Hb_000357_070 |
0.1059544481 |
- |
- |
- |
| 3 |
Hb_004678_010 |
0.107731271 |
transcription factor |
TF Family: Orphans |
Salt-tolerance protein, putative [Ricinus communis] |
| 4 |
Hb_017098_040 |
0.1100114995 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 5 |
Hb_002026_030 |
0.1113737483 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: mitogen-activated protein kinase kinase kinase YODA [Jatropha curcas] |
| 6 |
Hb_007765_130 |
0.1114489789 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 7 |
Hb_003055_070 |
0.116731641 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_112064_010 |
0.1170093768 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 9 |
Hb_003912_080 |
0.1174751733 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 10 |
Hb_034579_020 |
0.1206088068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001775_120 |
0.1220032706 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |
| 12 |
Hb_002740_100 |
0.1242345591 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 13 |
Hb_025045_010 |
0.1273106091 |
- |
- |
catalytic, putative [Ricinus communis] |
| 14 |
Hb_093077_010 |
0.1275705919 |
transcription factor |
TF Family: G2-like |
PREDICTED: two-component response regulator-like APRR2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_009449_100 |
0.1281257661 |
- |
- |
PREDICTED: equilibrative nucleotide transporter 1 [Jatropha curcas] |
| 16 |
Hb_005724_040 |
0.1289007908 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 17 |
Hb_001153_100 |
0.1289955221 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 18 |
Hb_002675_080 |
0.1291263778 |
- |
- |
PREDICTED: uncharacterized protein LOC105634950 [Jatropha curcas] |
| 19 |
Hb_004330_020 |
0.1294465528 |
- |
- |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 20 |
Hb_031385_010 |
0.1310938327 |
- |
- |
PREDICTED: alpha/beta hydrolase domain-containing protein 11 [Jatropha curcas] |