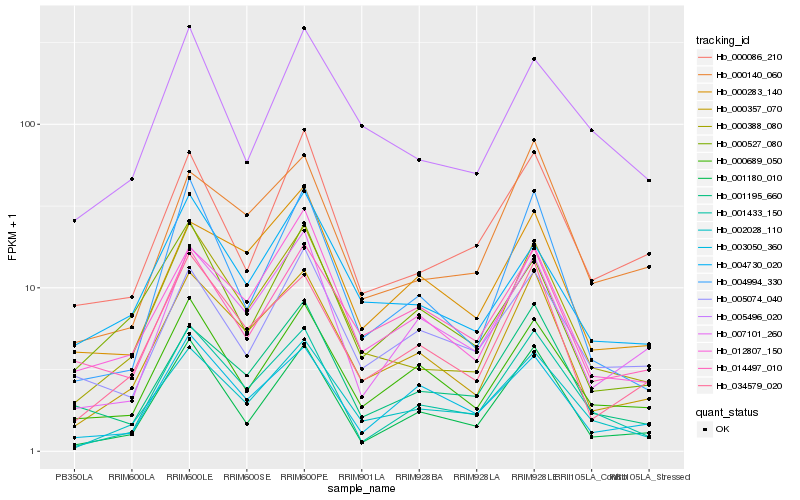
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002028_110 |
0.0 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 2 |
Hb_000357_070 |
0.1006912029 |
- |
- |
- |
| 3 |
Hb_000388_080 |
0.1078962889 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 4 |
Hb_001433_150 |
0.1097148979 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000689_050 |
0.1191208743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_012807_150 |
0.1219737059 |
- |
- |
PREDICTED: alpha-L-fucosidase 1 [Jatropha curcas] |
| 7 |
Hb_007101_260 |
0.1308761146 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 8 |
Hb_034579_020 |
0.1309390424 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000283_140 |
0.1316463542 |
- |
- |
PREDICTED: transcriptional corepressor LEUNIG-like isoform X1 [Jatropha curcas] |
| 10 |
Hb_005074_040 |
0.1322288399 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_005496_020 |
0.1322724173 |
- |
- |
PREDICTED: uncharacterized protein LOC105630867 [Jatropha curcas] |
| 12 |
Hb_003050_360 |
0.1349279892 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001195_660 |
0.1351215152 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 14 |
Hb_004994_330 |
0.1362815912 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 15 |
Hb_000086_210 |
0.1365059185 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 16 |
Hb_014497_010 |
0.1386533579 |
- |
- |
PREDICTED: uncharacterized protein LOC105635153 [Jatropha curcas] |
| 17 |
Hb_001180_010 |
0.1390245934 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 18 |
Hb_004730_020 |
0.1414400709 |
- |
- |
PREDICTED: uncharacterized protein LOC105641815 [Jatropha curcas] |
| 19 |
Hb_000140_060 |
0.1434251641 |
- |
- |
50S ribosomal protein L5, putative [Ricinus communis] |
| 20 |
Hb_000527_080 |
0.1445782833 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |