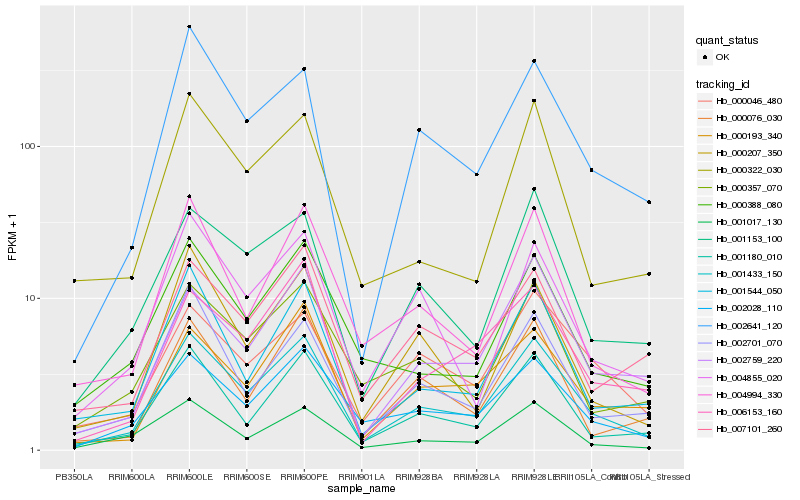
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001433_150 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001180_010 |
0.106198728 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 9 [Jatropha curcas] |
| 3 |
Hb_002028_110 |
0.1097148979 |
- |
- |
PREDICTED: F-box only protein 13 [Jatropha curcas] |
| 4 |
Hb_001153_100 |
0.1155774473 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Jatropha curcas] |
| 5 |
Hb_002641_120 |
0.118670495 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 6 |
Hb_001544_050 |
0.1193856638 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 7 |
Hb_002759_220 |
0.120187997 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 8 |
Hb_004994_330 |
0.1205566591 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_000357_070 |
0.1276069995 |
- |
- |
- |
| 10 |
Hb_000388_080 |
0.1285582673 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 11 |
Hb_000207_350 |
0.1291339755 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 12 |
Hb_000193_340 |
0.1297714317 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004855_020 |
0.1298925684 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 14 |
Hb_001017_130 |
0.1298931826 |
- |
- |
hypothetical protein POPTR_0010s06070g [Populus trichocarpa] |
| 15 |
Hb_002701_070 |
0.1304804694 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 16 |
Hb_000046_480 |
0.1312646266 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 5 [Jatropha curcas] |
| 17 |
Hb_007101_260 |
0.1326477675 |
- |
- |
altered response to gravity (arg1), plant, putative [Ricinus communis] |
| 18 |
Hb_000076_030 |
0.1330524486 |
- |
- |
PREDICTED: microtubule-associated protein TORTIFOLIA1 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000322_030 |
0.1342379194 |
- |
- |
PREDICTED: uncharacterized protein LOC105637911 [Jatropha curcas] |
| 20 |
Hb_006153_160 |
0.1348337972 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 2A isoform X4 [Jatropha curcas] |