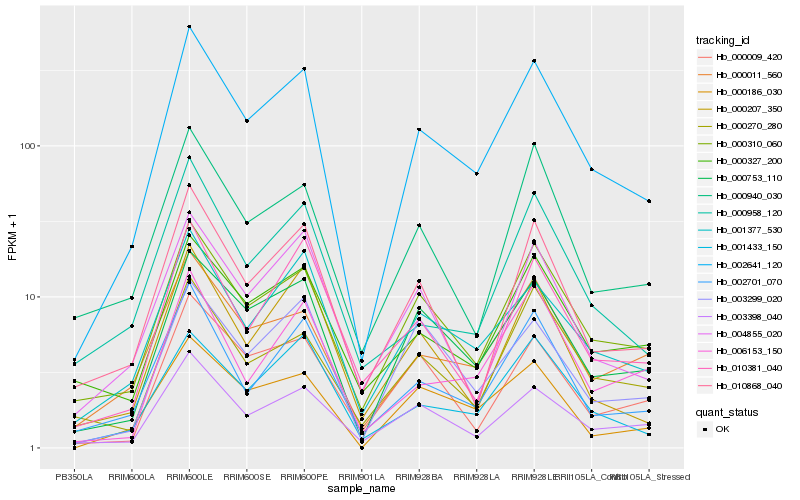
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002641_120 |
0.0 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 2 |
Hb_000753_110 |
0.0939563711 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X6 [Jatropha curcas] |
| 3 |
Hb_000310_060 |
0.0972288179 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 4 |
Hb_001377_530 |
0.1090027 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 5 |
Hb_003398_040 |
0.1169250057 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 6 |
Hb_002701_070 |
0.1178875399 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 7 |
Hb_001433_150 |
0.118670495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000186_030 |
0.1188732508 |
- |
- |
PREDICTED: uncharacterized protein LOC105649276 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000011_560 |
0.1225911611 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_010868_040 |
0.1266797799 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 11 |
Hb_000327_200 |
0.132941084 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 12 |
Hb_004855_020 |
0.1337454855 |
- |
- |
hypothetical protein POPTR_0006s10010g [Populus trichocarpa] |
| 13 |
Hb_003299_020 |
0.1354694435 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 14 |
Hb_000207_350 |
0.1363902006 |
- |
- |
PREDICTED: embryogenesis-associated protein EMB8 [Jatropha curcas] |
| 15 |
Hb_000940_030 |
0.137671193 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 16 |
Hb_010381_040 |
0.1391212253 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 17 |
Hb_000270_280 |
0.1399073388 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 18 |
Hb_000958_120 |
0.1399482817 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 19 |
Hb_006153_150 |
0.1409430051 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 20 |
Hb_000009_420 |
0.1411131751 |
- |
- |
PREDICTED: uncharacterized protein LOC105639614 isoform X2 [Jatropha curcas] |