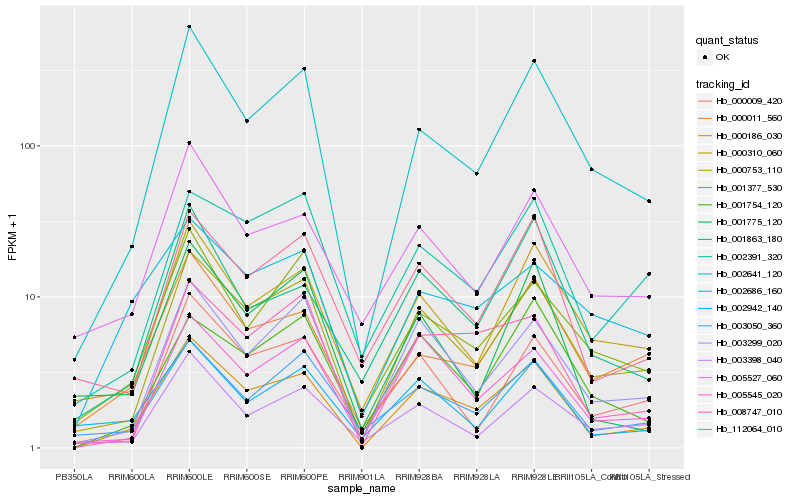
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000186_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649276 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000753_110 |
0.109636223 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BIM1 isoform X6 [Jatropha curcas] |
| 3 |
Hb_002641_120 |
0.1188732508 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 4 |
Hb_003299_020 |
0.1417485501 |
- |
- |
PREDICTED: uncharacterized protein LOC105649783 [Jatropha curcas] |
| 5 |
Hb_008747_010 |
0.146693258 |
- |
- |
PREDICTED: protein argonaute 2-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_000011_560 |
0.1499865788 |
- |
- |
PREDICTED: phosphoinositide phosphatase SAC2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000310_060 |
0.1502177141 |
- |
- |
hypothetical protein JCGZ_20793 [Jatropha curcas] |
| 8 |
Hb_005545_020 |
0.1542698622 |
- |
- |
endo beta n-acetylglucosaminidase, putative [Ricinus communis] |
| 9 |
Hb_112064_010 |
0.1569565892 |
- |
- |
PREDICTED: inositol-phosphate phosphatase [Jatropha curcas] |
| 10 |
Hb_003050_360 |
0.1598125405 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005527_060 |
0.1613621043 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 12 |
Hb_001377_530 |
0.1631024357 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 13 |
Hb_001863_180 |
0.1634872812 |
- |
- |
PREDICTED: serine/threonine-protein kinase SAPK2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000009_420 |
0.1683405465 |
- |
- |
PREDICTED: uncharacterized protein LOC105639614 isoform X2 [Jatropha curcas] |
| 15 |
Hb_003398_040 |
0.168798002 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002942_140 |
0.1695813463 |
- |
- |
PREDICTED: dual specificity protein phosphatase PHS1 [Jatropha curcas] |
| 17 |
Hb_002391_320 |
0.171757551 |
- |
- |
PREDICTED: phospholipase D delta-like [Jatropha curcas] |
| 18 |
Hb_001754_120 |
0.1734643568 |
- |
- |
PREDICTED: probable protein phosphatase 2C 40 [Jatropha curcas] |
| 19 |
Hb_002686_160 |
0.1747772777 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Populus euphratica] |
| 20 |
Hb_001775_120 |
0.175485281 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 12 [Jatropha curcas] |