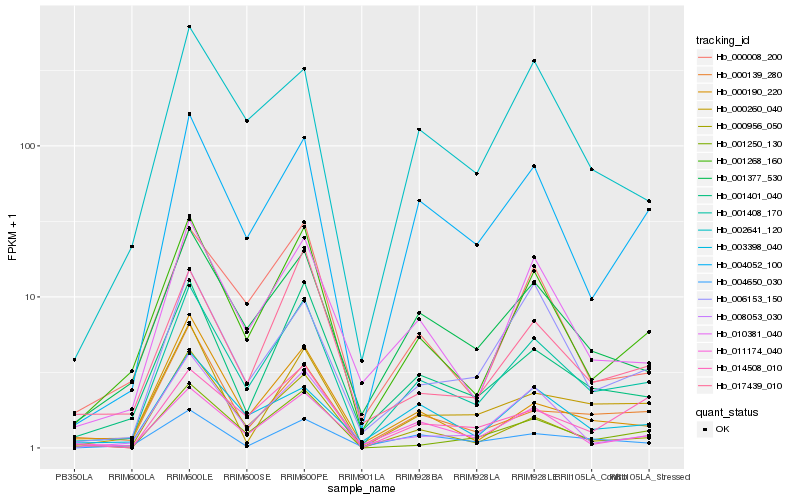
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001408_170 |
0.0 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 2 |
Hb_004052_100 |
0.1043977904 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 3 |
Hb_001268_160 |
0.1079074444 |
- |
- |
Glycerophosphodiester phosphodiesterase [Medicago truncatula] |
| 4 |
Hb_001401_040 |
0.1189009926 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 5 |
Hb_001377_530 |
0.1216489562 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 6 |
Hb_010381_040 |
0.1316984387 |
- |
- |
beta-galactosidase, putative [Ricinus communis] |
| 7 |
Hb_011174_040 |
0.139404402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001250_130 |
0.1493324333 |
- |
- |
hypothetical protein POPTR_0008s19280g [Populus trichocarpa] |
| 9 |
Hb_002641_120 |
0.1503898815 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 10 |
Hb_017439_010 |
0.1528599039 |
- |
- |
putative actin-97 protein [Linum usitatissimum] |
| 11 |
Hb_003398_040 |
0.153041075 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000008_200 |
0.1562942245 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 11 [Jatropha curcas] |
| 13 |
Hb_000260_040 |
0.156302542 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 14 |
Hb_006153_150 |
0.1563769735 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 15 |
Hb_000139_280 |
0.1592445091 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 16 |
Hb_008053_030 |
0.1594584799 |
- |
- |
Rop guanine nucleotide exchange factor, putative [Ricinus communis] |
| 17 |
Hb_004650_030 |
0.1596367111 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 18 |
Hb_014508_010 |
0.1622488965 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 19 |
Hb_000190_220 |
0.1637680409 |
- |
- |
PREDICTED: serine/threonine-protein kinase Nek2 [Jatropha curcas] |
| 20 |
Hb_000956_050 |
0.1644287034 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |