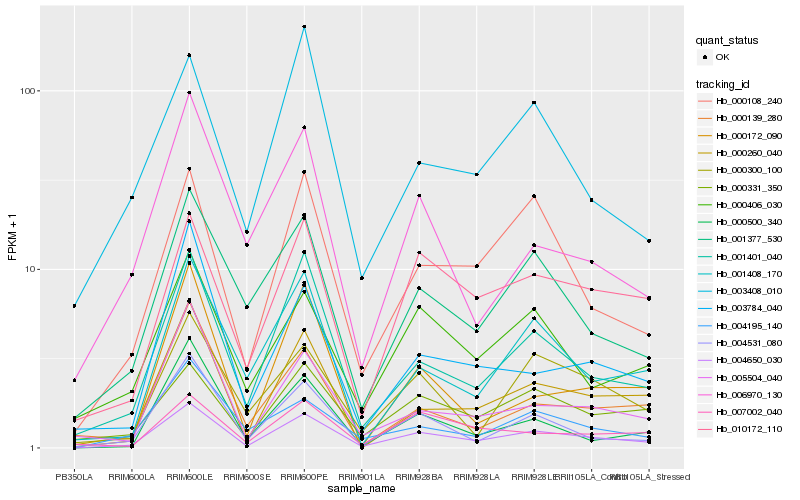
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004650_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 2 |
Hb_001401_040 |
0.1196193574 |
- |
- |
PREDICTED: protein CYPRO4 [Jatropha curcas] |
| 3 |
Hb_000108_240 |
0.1416312514 |
- |
- |
hypothetical protein Csa_1G600150 [Cucumis sativus] |
| 4 |
Hb_000260_040 |
0.1431495678 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 5 |
Hb_004531_080 |
0.1440630634 |
- |
- |
hypothetical protein RCOM_1491960 [Ricinus communis] |
| 6 |
Hb_001377_530 |
0.1512385246 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 7 |
Hb_010172_110 |
0.1548485491 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 8 |
Hb_001408_170 |
0.1596367111 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 2 [Jatropha curcas] |
| 9 |
Hb_000331_350 |
0.1600167481 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 10 |
Hb_000406_030 |
0.163745879 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 11 |
Hb_004195_140 |
0.1640662927 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 12 |
Hb_000300_100 |
0.1641735383 |
- |
- |
NADH-cytochrome b5 reductase 1 [Morus notabilis] |
| 13 |
Hb_005504_040 |
0.1662979554 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003784_040 |
0.171300431 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000500_340 |
0.1736894271 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 16 |
Hb_000172_090 |
0.1779248176 |
- |
- |
PREDICTED: uncharacterized protein LOC105650755 [Jatropha curcas] |
| 17 |
Hb_007002_040 |
0.1793843021 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 18 |
Hb_003408_010 |
0.1802569903 |
- |
- |
fructokinase [Manihot esculenta] |
| 19 |
Hb_006970_130 |
0.1806651577 |
- |
- |
PREDICTED: histone H3.3-like [Colobus angolensis palliatus] |
| 20 |
Hb_000139_280 |
0.1813391951 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |