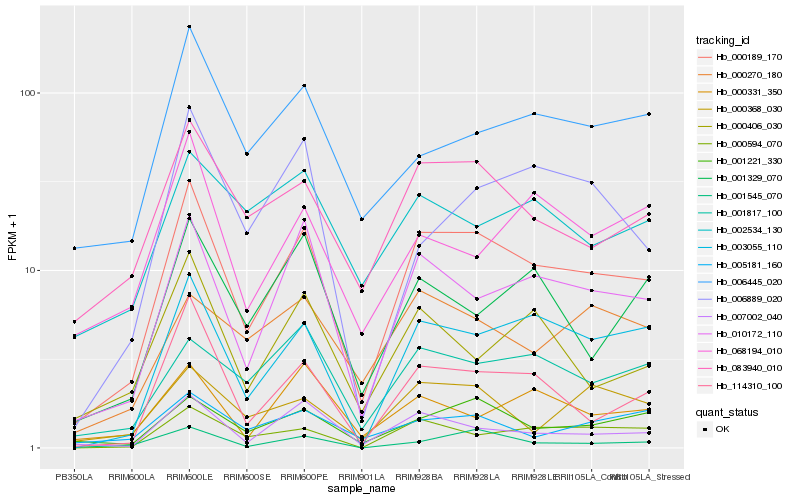
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000189_170 |
0.0 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_010172_110 |
0.1265814954 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 3 |
Hb_003055_110 |
0.137135242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_083940_010 |
0.1448113927 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 5 |
Hb_007002_040 |
0.1507104127 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 6 |
Hb_114310_100 |
0.1524105339 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 7 |
Hb_001221_330 |
0.1558835525 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 8 |
Hb_000594_070 |
0.1684547695 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_001817_100 |
0.1695171004 |
- |
- |
PREDICTED: deoxyhypusine hydroxylase [Jatropha curcas] |
| 10 |
Hb_001545_070 |
0.1758600265 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000406_030 |
0.1796147924 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 12 |
Hb_006445_020 |
0.1798523143 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 13 |
Hb_001329_070 |
0.1820362139 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 14 |
Hb_000368_030 |
0.1821840412 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 15 |
Hb_006889_020 |
0.186385086 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 16 |
Hb_000331_350 |
0.1869734124 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 17 |
Hb_005181_160 |
0.1890120259 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 18 |
Hb_002534_130 |
0.1899856587 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 19 |
Hb_000270_180 |
0.191708324 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 20 |
Hb_068194_010 |
0.1933252544 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |