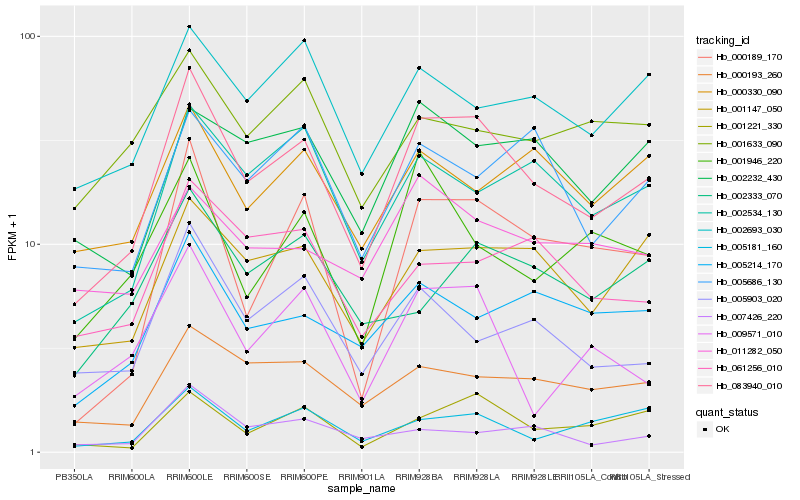
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083940_010 |
0.0 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 2 |
Hb_001147_050 |
0.1368307575 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase Cx32, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000193_260 |
0.1406064953 |
- |
- |
PREDICTED: protein NEN1 [Jatropha curcas] |
| 4 |
Hb_000189_170 |
0.1448113927 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 5 |
Hb_002534_130 |
0.1453666365 |
- |
- |
membrane associated ring finger 1,8, putative [Ricinus communis] |
| 6 |
Hb_005214_170 |
0.1521152807 |
- |
- |
PREDICTED: uncharacterized protein LOC105636021 [Jatropha curcas] |
| 7 |
Hb_001221_330 |
0.152238815 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 8 |
Hb_002693_030 |
0.1538666663 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 9 |
Hb_005181_160 |
0.1564154788 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 10 |
Hb_005903_020 |
0.1578483558 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 11 |
Hb_002232_430 |
0.1590040644 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 12 |
Hb_061256_010 |
0.162503476 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 13 |
Hb_009571_010 |
0.163551828 |
- |
- |
early-responsive to dehydration family protein [Populus trichocarpa] |
| 14 |
Hb_000330_090 |
0.1645838597 |
- |
- |
ornithine carbamoyltransferase, putative [Ricinus communis] |
| 15 |
Hb_005686_130 |
0.1660991296 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
isopentenyl pyrophosphate isomerase [Hevea brasiliensis] |
| 16 |
Hb_002333_070 |
0.1664785918 |
- |
- |
PREDICTED: ACT domain-containing protein ACR4-like [Jatropha curcas] |
| 17 |
Hb_011282_050 |
0.1709243212 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_001633_090 |
0.1726054381 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 19 |
Hb_001946_220 |
0.1727414353 |
- |
- |
PREDICTED: signal peptidase complex catalytic subunit SEC11A-like [Jatropha curcas] |
| 20 |
Hb_007426_220 |
0.172747553 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |