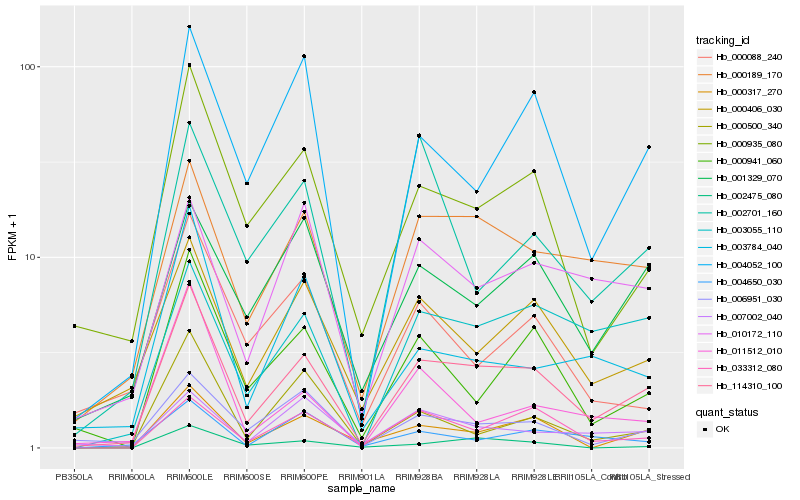
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114310_100 |
0.0 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 2 |
Hb_000189_170 |
0.1524105339 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_000935_080 |
0.1617768335 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000406_030 |
0.1716062896 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 5 |
Hb_000317_270 |
0.1719240368 |
- |
- |
Uncharacterized protein TCM_001673 [Theobroma cacao] |
| 6 |
Hb_000941_060 |
0.1767649375 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0016s05890g [Populus trichocarpa] |
| 7 |
Hb_004052_100 |
0.1787117786 |
- |
- |
PREDICTED: protein SPIRAL1-like 3 [Jatropha curcas] |
| 8 |
Hb_006951_030 |
0.1829059912 |
- |
- |
PREDICTED: protein MCM10 homolog [Jatropha curcas] |
| 9 |
Hb_003055_110 |
0.1876317333 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_033312_080 |
0.1895682766 |
- |
- |
aldose-1-epimerase, putative [Ricinus communis] |
| 11 |
Hb_001329_070 |
0.1904170933 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 12 |
Hb_007002_040 |
0.1934129334 |
transcription factor |
TF Family: PHD |
DNA replication regulator dpb11, putative [Ricinus communis] |
| 13 |
Hb_010172_110 |
0.1997449213 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 14 |
Hb_003784_040 |
0.2039454101 |
- |
- |
Afadin- and alpha-actinin-binding protein, putative [Ricinus communis] |
| 15 |
Hb_004650_030 |
0.2050037936 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 16 |
Hb_002475_080 |
0.2071522461 |
transcription factor |
TF Family: PHD |
PREDICTED: PHD finger protein MALE MEIOCYTE DEATH 1 [Jatropha curcas] |
| 17 |
Hb_000500_340 |
0.2098054481 |
- |
- |
PREDICTED: exonuclease 1 [Jatropha curcas] |
| 18 |
Hb_011512_010 |
0.210841487 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 19 |
Hb_000088_240 |
0.2111627579 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 20 |
Hb_002701_160 |
0.2129945862 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 1 [Jatropha curcas] |