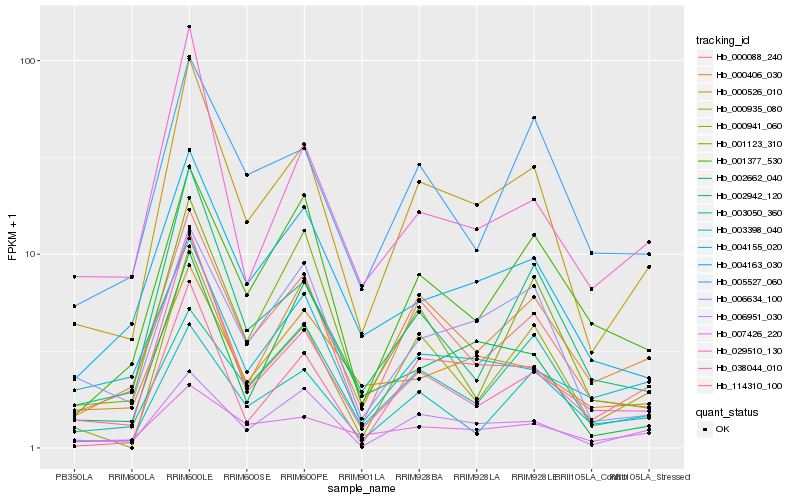
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000935_080 |
0.0 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_000088_240 |
0.1057749897 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 3 |
Hb_000941_060 |
0.128296198 |
transcription factor |
TF Family: HB |
hypothetical protein POPTR_0016s05890g [Populus trichocarpa] |
| 4 |
Hb_006951_030 |
0.1314333898 |
- |
- |
PREDICTED: protein MCM10 homolog [Jatropha curcas] |
| 5 |
Hb_006634_100 |
0.1377469098 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002942_120 |
0.1442225615 |
transcription factor |
TF Family: bHLH |
Transcription factor ICE1, putative [Ricinus communis] |
| 7 |
Hb_004163_030 |
0.1443628868 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 8 |
Hb_005527_060 |
0.1493627783 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 9 |
Hb_000406_030 |
0.1539550406 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 10 |
Hb_038044_010 |
0.1548863567 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 11 |
Hb_003398_040 |
0.1576967711 |
- |
- |
PREDICTED: actin-related protein 5 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001377_530 |
0.1607272891 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 13 |
Hb_114310_100 |
0.1617768335 |
- |
- |
glucose-1-phosphate adenylyltransferase, putative [Ricinus communis] |
| 14 |
Hb_007426_220 |
0.1637574621 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 15 |
Hb_004155_020 |
0.1663879719 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 16 |
Hb_029510_130 |
0.1667965723 |
- |
- |
PREDICTED: malonate--CoA ligase [Jatropha curcas] |
| 17 |
Hb_003050_360 |
0.1668390466 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002662_040 |
0.1669199515 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 19 |
Hb_000526_010 |
0.1690762616 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 20 |
Hb_001123_310 |
0.1692701665 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |