| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
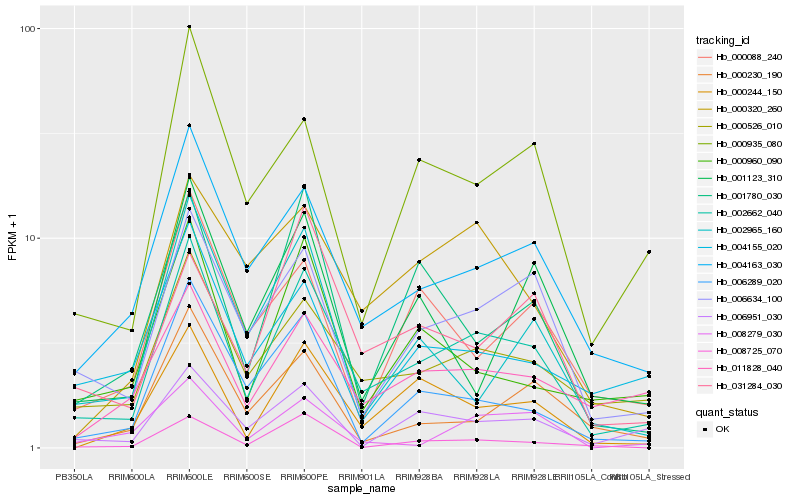
Hb_002662_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 2 |
Hb_000526_010 |
0.1377324521 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 3 |
Hb_004163_030 |
0.1391279559 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 4 |
Hb_006634_100 |
0.1493556946 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000244_150 |
0.1656022358 |
- |
- |
PREDICTED: chromatin assembly factor 1 subunit FAS2 [Jatropha curcas] |
| 6 |
Hb_000935_080 |
0.1669199515 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_011828_040 |
0.1752354152 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 8 |
Hb_006289_020 |
0.1780307981 |
- |
- |
PREDICTED: shugoshin-1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_006951_030 |
0.1789357665 |
- |
- |
PREDICTED: protein MCM10 homolog [Jatropha curcas] |
| 10 |
Hb_000960_090 |
0.1810692911 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001780_030 |
0.1826123094 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At5g41260 [Jatropha curcas] |
| 12 |
Hb_008725_070 |
0.1864984471 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 13 |
Hb_004155_020 |
0.1921213586 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 14 |
Hb_000320_260 |
0.1921742329 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000088_240 |
0.193661228 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_031284_030 |
0.1939594609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000230_190 |
0.1944419692 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_002965_160 |
0.1952667541 |
- |
- |
PREDICTED: uncharacterized protein LOC105643503 [Jatropha curcas] |
| 19 |
Hb_008279_030 |
0.197791724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001123_310 |
0.1985879695 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |